FY2014 Annual Report

Abstract
The genome contains all the genetic information of a given organism. Decoding the genome provides the molecular basis for understanding every biological phenomenon. Since the turn of the 21st century, genomes of various metazoans have been sequenced, and consequently studies progressed efficiently in the fields of evolutionary biology, developmental biology, and environmental biology. The objective of the Marine Genomic Unit (MGU) is to decode the genomes of target marine organisms, especially marine invertebrates, so as to comprehensively elucidate the molecular mechanisms underlying (a) development and evolution of marine invertebrates, (b) environmental responses of marine organisms, and (c) specific functions of metazoans. In fiscal year 2014, we obtained several results of studies in the three research areas each with publication of approximately 20 reports.
(a) Developmental and evolutionary genomics:
We are interested in the origin and evolution of chordates. We have already analyzed an evolutionary scenario within three groups of chordates. Since chordates originated from common ancestor shared with non-chordate deuterostomes such as echinoderms and hemichordates, a big question remained is an evolutionary link between non-chordate deuterostomes and chordates. This year, we have discussed the origins and evolution of chordates and proposed the three-phylum system of chordates.
(b) Environmental genomics:
The coral reefs of the Okinawa islands are amongst the most biologically diverse ecosystems in the world. The key organisms in their establishment, the scleractinian corals, increasingly face a range of human-caused challenges including seawater temperature rises and ocean acidification. To understand better the molecular mechanisms underlying coral biology, we succeeded in deciphering the 420-Mbp-long genome of the coral Acropora digitifera in 2011, and found that the coral innate immunity repertoire is notably more complex than that of the sea anemone. In addition, in 2013, we succeeded in deciphering the 1500-Mbp-long genome of a zooxanthella, Symbiodinium minutum. This year, Chuya Shinzato developed a novel, cross-species microsatellite markers for Acropora corals using next-generation sequencing technology.
(c) Functional genomics:
We continue to decode genomes of marine invertebrates in relation to functional genomics. This year, in collaboration with Dr. Kikawada’s group at the National Institute of Agrobiological Sciences (NIAS), Tsukuba, Japan, we have sequenced the genome of an anhydrobiotic midge with an extreme desiccation tolerance ability.
1. Staff
- Group Leaders
- Takeshi Kawashima
- Eiichi Shoguchi
- Chuya Shinzato
- Staff Scientists
- Mayuko Hamada
- Keisuke Nakashima
- Konstantin Khalturin
- Postdoctoral Scholars
- Ken Maeda
- Takeshi Takeuchi
- Fuki Gyoja
- Yuuri Yasuoka (JSPS, PD Fellow)
- Koki Nishitsuji
- Yuna Zayasu
- Students
- Kenneth Baughman
- Keita Ikegami
- Yi-Jyun Luo
- Tsai-Ming Lu
- Technical Staffs
- Kanako Hisata
- Sakura Kikuchi
- Miyuki Kanda
- Mariia Khalturina
- Yoshie Nishitsuji
- Research Assistants
- Yuki Yasuoka
- Research Administrators
- Shoko Yamakawa
- Tomomi Teruya
2. Collaborations
- Theme: Anhydrobiotic midge
- Type of collaboration: Collaboration
- Researchers:
- Dr. Takahiro Kikawada, National Institute of Agrobiological Sciences (NIAS), Tsukuba, Japan
- Theme: Chip-sequence analysis of Ci-Bra target genes
- Type of collaboration: Collaboration
- Researchers:
- Dr. Hiroki Takahashi, National Institute for Basic Biology, Japan
- Theme: Analysis of Ciona genes with transgenic lines
- Type of collaboration: Collaboration
- Researchers:
- Dr. Yasunori Sasakura, Shimoda Marine Research Center, University of Tsukuba, Japan
- There are several collaborative works not specifically listed here.
3. Activities and Findings
3.1 Developmental and evolutionary genomics
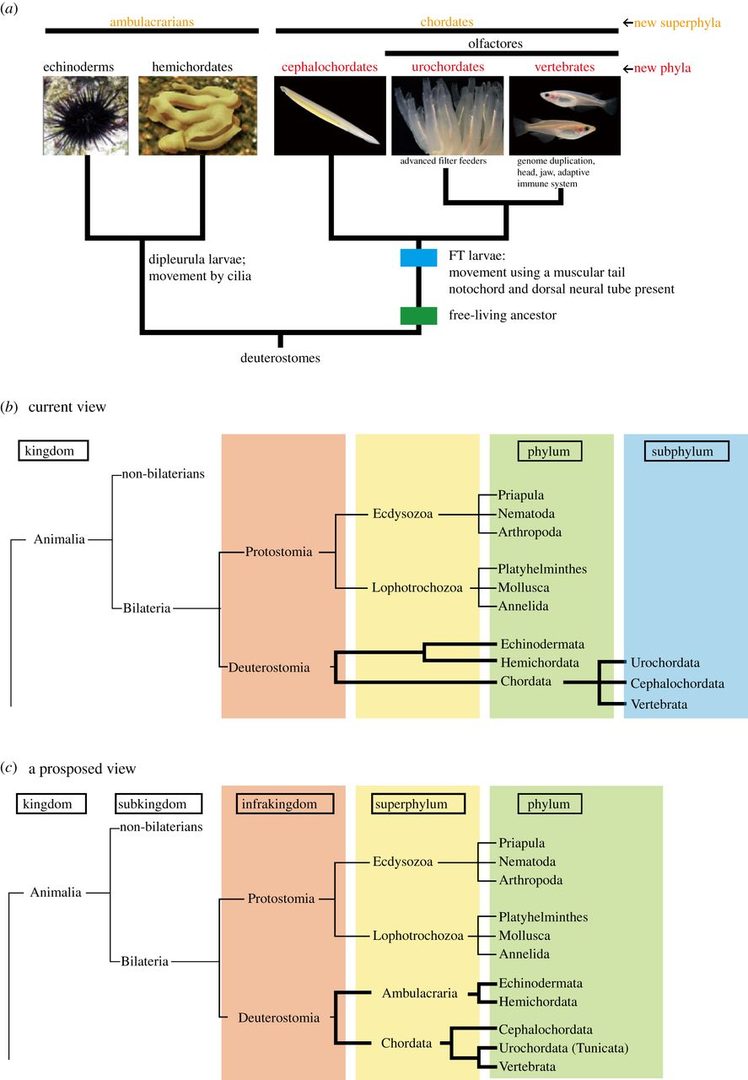
We are interested in the origin and evolution of chordates. This year, we have discussed the origins and evolution of chordates and proposed the three-phylum system of chordates. Traditional metazoan phylogeny classifies the Vertebrata as a subphylum of the phylum Chordata, together with two other subphyla, the Urochordata (Tunicata) and the Cephalochordata. The Chordata, together with the phyla Echinodermata and Hemichordata, comprise a major group, the Deuterostomia. Chordates invariably possess a notochord and a dorsal neural tube. Although the origin and evolution of chordates has been studied for more than a century, few authors have intimately discussed taxonomic ranking of the three chordate groups themselves. Accumulating evidence shows that echinoderms and hemichordates form a clade (the Ambulacraria), and that within the Chordata, cephalochordates diverged first, with tunicates and vertebrates forming a sister group. Chordates share tadpole-type larvae containing a notochord and hollow neural tube, whereas ambulacrarians have dipleurula-type larvae containing a hydrocoel. We have proposed that an evolutionary occurrence of tadpole-type larvae is fundamental to understanding mechanisms of chordate origin. Protostomes have now been reclassified into two major taxa, the Ecdysozoa and Lophotrochozoa, whose developmental pathways are characterized by ecdysis and trochophore larvae, respectively. Consistent with this classification, the profound dipleurula versus tadpole larval differences merit a category higher than the phylum. Thus, it is recommended that the Ecdysozoa, Lophotrochozoa, Ambulacraria and Chordata be classified at the superphylum level, with the Chordata further subdivided into three phyla, on the basis of their distinctive characteristics (Figure 1).
3.2 Environmental genomics
The genus Acropora (Scleractinia, Acroporidae) is one of the most widespread coral genera, comprising the largest number of extant species among scleractinian (reef-building) corals. Molecular phylogenetic studies have suggested that A. tenuis belongs to the most basal clade (clade I) while A. digitifera belongs to a derived clade (clade IV). In order to develop microsatellite markers that would be useful for most Acropora species, we sequenced the genomic DNA of A. tenuis using a next generation sequencer (Illumina MiSeq), and designed primer sets that amplify microsatellite loci. Afterward we selected primer pairs with perfectly matched nucleotide sequences from which at least one primer was uniquely mapped to the A. digitifera genome. Fourteen microsatellite markers showed non-significant departure from Hardy–Weinberg equilibrium(HWE) in both A. tenuis and A. digitifera. Thus these markers could be used for wide range of species and may provide powerful tools for population genetics studies and conservation of Acropora corals.
3.3 Functional genomics
This year, in collaboration with Dr. Kikawada’s group at the National Institute of Agrobiological Sciences (NIAS), Tsukuba, Japan, we have sequenced the genome of an anhydrobiotic midge with an extreme desiccation tolerance ability. Anhydrobiosis represents an extreme example of tolerance adaptation to water loss, where an organism can survive in an ametabolic state until water returns. We reported the first comparative analysis examining the genomic background of extreme desiccation tolerance, which is exclusively found in larvae of the only anhydrobiotic insect, Polypedilum vanderplanki. We compare the genomes of P. vanderplanki and a congeneric desiccation-sensitive midge P. nubifer. We determine that the genome of the anhydrobiotic species specifically contains clusters of multi-copy genes with products that act as molecular shields (Figure 2). In addition, the genome possesses several groups of genes with high similarity to known protective proteins. However, these genes are located in distinct paralogous clusters in the genome apart from the classical orthologues of the corresponding genes shared by both chironomids and other insects. The transcripts of these clustered paralogues contribute to a large majority of the mRNA pool in the desiccating larvae and most likely define successful anhydrobiosis. Comparison of expression patterns of orthologues between two chironomid species provides evidence for the existence of desiccation-specific gene expression systems in P. vanderplanki.

ARIds are genomic regions containing clusters of duplicated genes that are transcriptionally active during anhydrobiosis. (a) A gene of foreign origin (for example, LEA protein) is incorporated into P. vanderplanki genome by HGT and undergoes extensive duplications and shuffling. (b) A pre-existing P. vanderplanki gene originally not involved in anhydrobiosis and originating from another region of P. vanderplanki genome was inserted to a new locus by intragenomic duplication (IGD) and undergoes extensive duplications and shuffling to acquire or improve a specific function for desiccation tolerance. All the genes in the ARIds from both a,b become highly upregulated during anhydrobiosis (red arrows).
4. Publications
4.1 Journals
- Satoh, N., Rokhsar, D., Nishikawa, T.
Chordate evolution and the three-phylum system.
Proc. R. Soc. B. 281:1794 (2014)
- Yasuoka, Y., Suzuki, Y., Takahashi S., Someya, H., Sudou, N., Haramoto, Y., Cho, K.W., Asashima, M., Sugano, S., Taira, M.
Occupancy of tissue-specific cis-regulatory modules by Otx2 and TLE/Groucho for embryonic head specification.
Nature communications, DOI: 10.1038/ncomms5322. (2014)

- Iitsuka T, Mita K, Hozumi A, Hamada M, Satoh N, Sasakura Y.
Transposon-mediated targeted and specific knockdown of maternally expressed transcripts in the ascidian Ciona intestinalis.
Sci Rep. doi: 10.1038/srep05050. (2014)
- Gyoja F.
A genome-wide survey of bHLH transcription factors in the Placozoan Trichoplax adhaerens reveals the ancient repertoire of this gene family in metazoan..
Gene 542:29-37. (2014)
- Tagawa K, Arimoto A, Sasaki A, Izumi M, Humphreys T, Fujiyama A, Kagoshima H, Shin-I T, Kohara Y, Satoh N, Kawashima T.
A cDNA resource for gene expression studies of a hemichordate, Ptychodera flava.
Zool. Sci., 31(7):414-20. (2014)
- Ebchuqin E, Yokota N, Yamada L, Yasuoka Y, Akasaka M, Arakawa M, Deguchi R, Mori T, Sawada H.
Evidence for participation of GCS1 in fertilization of the starlet sea anemone Nematostella vectensis: Implication of a common mechanism of sperm-egg fusion in plants and animals.
Biochem Biophys Res Commun. 5;451(4):522-528. doi: 10.1016/j.bbrc.2014.08.006. (2014)
- Satou Y, Hirayama K, Mita K, Fujie M, Chiba S, Yoshida R, Endo T, Sasakura Y, Inaba K, Satoh N.
Sustained heterozygosity across a self-incompatibility locus in an inbred ascidian.
Mol Biol Evol. on line. (2014)
- Baughman KW, McDougall C, Cummins SF, Hall M, Degnan BM, Satoh N, Shoguchi E.
Genomic organization of Hox and ParaHox clusters in the echinoderm, Acanthaster planci.
Genesis. , 52(12): 952-8. (2014)
- Satoh N, Tagawa K, Lowe CJ, Yu JK, Kawashima T, Takahashi H, Ogasawara M, Kirschner M, Hisata K, Su YH, Gerhart J
On a possible evolutionary link of the stomochord of hemichordates to pharyngeal organs of chordates
genesis, 52(12): 925–934 (2014)
- Shinzato, C., Yasuoka, Y., Mungpakdee, D., Arakaki, N., Fujie, M., Nakajima, Y., Satoh, N.
Development of novel, cross-species microsatellite markers for Acropora corals using next-generation sequencing technology.
Front. Mar. Sci., doi: 10.3389/fmars.2014.00011 (2014)
- Mungpakdee, S., Shinzato, C., Takeuchi, T., Kawashima, T., Koyanagi, R., Hisata, K., Tanaka, M., Goto, H., Fujie, M., Lin, S., Satoh, N., and Shoguchi, E.
Massive gene transfer and extensive RNA editing of a symbiotic dinoflagellate plastid genome.
Genome Biol Evol. 6(6):1408-22. doi: 10.1093/gbe/evu109. (2014)
- Shinzato, C., Mungpakdee, S., Satoh, N., Shoguchi, E.
A genomic approach to coral-dinoflagellate symbiosis: Studies of Acropora digitifera and Symbiodinium minutum. Frontier in Microbiology. doi: 10.3389/fmicb.2014.00336. (2014)

- Tsuta H, Shinzato C, Satoh N, Hidaka M.
Telomere Shortening in the Colonial Coral Acropora digitifera During Development.
Zoolog Sci 31(3):129-34. (2014)
- Nakajima Y., Shinzato, C., Inagaki, F., Satoh, N., Khalturina, M., Mitarai, S.
Cross-species, amplifiable microsatellite markers for neoverrucid barnacles from deep-sea hydrothermal vents developed using next-generation sequencing.
Int. J. Mol. Sci. 2014, 15, 14364-14371; doi:10.3390/ijms150814364 (2014)
- Bosch,T.C.G., Adamska, M., Augustin, R., Domazet-Loso, T., Foret, S., Fraune, S., Funayama, N., Grasis, J., Hamada, H., Hatta, M., Hobmayer, B., Kawai, K., Klimovich, A., Manuel, M., Shinzato, C., Technau, U., Yum, S., J. Miller, D.J.
How do environmental factors influence life cycles and development? An experimental framework for early-diverging metazoans.
Bioessays. 36, 1185-1194 (2014)
- Zayasu, Y., Miyazaki, K., Lien, Y.T., Okubo, N.
Direct evidence of sexual reproduction in the zebra coral, Oulastrea crispata (Anthozoa, Scleractinia), in Japan
Invertebrate Reproduction and Development 59: 61-65 (2015) DOI: 10.1080/07924259.2015.1006340
- Gusev O, Suetsugu Y, Cornette R, Kawashima T, Logacheva MD, Kondrashov AS, Penin AA, Hatanaka R, Kikuta S, Shimura S, Kanamori H, Katayose Y, Matsumoto T, Shagimardanova E, Alexeev D, Govorun V, Wisecaver J, Mikheyev A, Koyanagi R, Fujie M, Nishiyama T, Shigenobu S, Shibata TF, Golygina V, Hasebe M, Okuda T, Satoh N, Kikawada T.
Comparative genome sequencing reveals genomic signature of extreme desiccation tolerance in the anhydrobiotic midge.
Nat Commun. 12;5:4784. doi: 10.1038/ncomms5784. (2014)
- Maeda K.
51 species in “Gobioidei (pp 1215–1339)” Okiyama M. (ed) An Atlas of Early Stage Fishes in Japan Second Edition
Tokai University Press, Japan (2014) - Maeda K.
Stiphodon niraikanaiensis, a new species of sicydiine goby from Okinawa Island (Gobiidae: Sicydiinae).
Ichthyological Research, 61: 99-107 (2014)
- Maeda K, Tachihara K.
Larval fish fauna of a sandy beach and an estuary on Okinawa Island, focusing on larval habitat utilization by the suborder Gobioidei.
Fish Sci 80: 1215–1229 (2014)
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- Baughman, K., Satoh, N., and Shoguchi, E. Hox and ParaHox in star-fish Acanthaster planci. The 85th Annual Meeting of the Zoological Society of Japan. (Tohoku University, Kawauchi-Kita Campus, Sendai, Miyagi Pref.) (2014)
- Hamada, M. Genome analysis of the symbiotic system in Cnidarian. In Meeting of the Zoological Society of Japan Chubu Area Branch (Notokin Plaza Hotel, Noto, Ishikawa Pref.). (2014)
- Hamada, M., Satoh, N., and Bosch, T.C.G. Molecular interactions and genome evolution in symbiosis using green hydra. In The 85th Annual Meeting of the Zoological Society of Japan (Tohoku University, Kawauchi-Kita Campus, Sendai, Miyagi Pref.). (2014)
- Hamada, M., Shinzato, C., Kürn, U., Satoh, N., and Bosch, T.C.G. Analysis of molecular interactions and genome evolution in symbiosis using green hydra. In Meeting of Japanese Society of Phycology (Kyushu University (Fukuoka, Japan)). (2015)
- Iida, M., Kondo, M., Maeda, K., and Tachihara, K. Dispersal strategy of amphidromous gobioid larvae. In Symposium of the 2014 autumn meeting of the Japanese Society of Fisheries Science "Latest studies on early life history of fish" (Kyushu University, Fukuoka, Japan). (2014)
- Iida, M., Kondo, M., Maeda, K., and Tachihara, K. Early life history traits of Okinawan amphidromous gobioid fishes. In The first Okinawa fish researcher's meeting (Okinawa Prefecture Nago Youth Center). (2015)
- Ikegami, K., Hamada, M., and Satoh, N. Towards understanding of the phylotypic stage in the ascidian Ciona intestinalis. In The 85th Annual Meeting of the Zoological Society of Japan (Tohoku University, Kawauchi-Kita Campus, Sendai, Miyagi Pref.). (2014)
- Imoto, J., Miyashita, M., Gotoh, R., Okuizumi, K., Shinzato, C., Satoh, N., and Hanzawa, N. A comprehensive molecular phylogeny of Scyphozoa based on mitochondrial genome. In The 85th Annual Meeting of the Zoological Society of Japan (Tohoku University, Kawauchi-Kita Campus, Sendai, Miyagi Pref.). (2014)
- Kanda, M., Koyanagi, R., Fujie, M., Sakamoto, T., Sakamoto, H., and Satoh, N. Genome decoding of a phoronida Phoronis australis and a nemertea Lineus geniculatus. In The 85th Annual Meeting of the Zoological Society of Japan (Tohoku University, Kawauchi-Kita Campus, Sendai, Miyagi Pref.). (2014)
- Kawashima, T. Reconstruction of paleo genomic information of metazoan based on a microsyteny analysis. In Japan Geoscience Union Meeting 2014 (Pacifico Yokohama, Kanagawa). (2014)
- Kawashima, T. "Had there been no ECAT, now we would come up against ...". In The 17th meeting for Information exchange in the group grant for the Evolution of complex Adaptive Traits. (National Institute of Basic Biology, Okazaki, Aichi Pref.). (2014)
- Koyanagi, R., Kanda, M., Fujie, M., Suzuki, T., and Satoh, N. Genome decoding of a gastroricha, Ichthydium podura. In The 85th Annual Meeting of the Zoological Society of Japan (Tohoku University, Kawauchi-Kita Campus, Sendai, Miyagi Pref.). (2014)
- Luo, Y.-J., Takeuchi, T., Koyanagi, R., Tanaka, M., Khalturina, M., Fujie, M., Yamasaki, S., Yamada, L., Sawada, H., Endo, K., et al. The Lingula genome and the evolution of lophotrochozoans and biomineralization. The 85th Annual Meeting of the Zoological Society of Japan. (Tohoku University, Kawauchi-Kita Campus, Sendai, Miyagi Pref.) (Year)
- Maeda, K. Fish in Manko. In Special Talk about Fish in Manko (Manko Waterbird and Wetland Center). (2014)
- Maeda, K. Story of sicydiine goby study. In The first Okinawa fish researchers' meeting (Okinawa Prefecture Nago Youth Center). (2015)
- Maeda, K., Kondo, M., Saito, M., Iida, M., and Tachihara, K. Variation of amphidromy: examples from gobioids inhabiting Okinawan streams. In Symposium of the Ichthyological Society of Japan "Amphidromy in fishes: its life-history diversification and evolution". (Kanagawa Prefectural Museum of Natural History, Kanagawa, Japan). (2014)
- Maeda, K., Kondo, M., Saito, M., Iida, M., and Tachihara, K. What is amphidromy? Discussion with variety of migration patterns. In GORI Meeting 2014 (College of Bioresource Science, Nihon University (Fujisawa-shi, Kanagawa)). (2014)
- Maeda, K., and Tachihara, K. Recruitment mechanism of freshwater and estuarine gobies. In Symposium of the 2014 autumn meeting of the Japanese Society of Fisheries Science "Latest studies on early life history of fish" (Kyushu University, Fukuoka, Japan). (2014)
- Nakajima, Y., Shinzato, C., Zayasu, Y., Satoh, N., and Mitarai, T. The reproductive traits and genetic differentiations of among types and regions of Galaxea fascicularis in The Nansei Islands. In The 17th Annual Conference of the Japanese Coral Reef Society (Kochijo-Hall & Kochi Kaikan, Kochi, Japan). (2014)
- Satoh, N. Genome decoding project of marine invertebrates at OIST. SMBE 2014 - S5: Establishing a " Global Invertebrate Genome Alliance" (GIGA) for Comparative Genomics". (San Juan, Puerto Rico) (2014)
- Satoh, N. Chordate evolution and three-phylum system. In Science Council of Japan Symposium "Recent advance in evolutionary biology" (Science Council of Japan Auditorium). (2014)
- Satoh, N. Genome scientific basis of coral and Symbiodinium symbiosis. In APCRS (The 3rd Asia-Pacific Coral Reef Symposium) (Howard Hotel, Pingtung, Taiwan). (2014)
- Satou, Y., Hirayama, K., Mita, K., Fujie, M., Chiba, S., Yoshida, R., Endo, T., Sasakura, Y., Inaba, K., and Satoh, N. Sustained heterozygosity across a self-incompatibility locus in an inbred ascidian. In The 85th Annual Meeting of the Zoological Society of Japan (Tohoku University, Kawauchi-Kita Campus, Sendai, Miyagi Pref.). (2014)
- Shinzato, C. Decoding the genome of a non-model organism. Practical Workshop on High-Throughput Sequencing Data Analysis. (OIST) (2014)
- Shinzato, C. Frontiers of coral science. In Scientific lecture and Science Cafe, "Sango no Kimochi (What Corals Think), OIST Science Trip in Naha 2015 (Okinawa Prefecture Museum & Art Museum). (2015)
- Shinzato, C., Yasuoka, Y., Mungpakdee, S., Zayasu, Y., Arakaki, N., Fujie, M., Nakajima, Y., and Satoh, N. Development of novel, cross-species microsatellite markers for Acropora corals using next-generation sequencing technology. In The 85th Annual Meeting of the Zoological Society of Japan (Tohoku University, Kawauchi-Kita Campus, Sendai, Miyagi Pref). (2014)
- Shoguchi, E. Enjoyment and difficulty on decoding de novo genomes of marine organisms. In The 21st meeting for community of young biologists around the sea. (Shimane University (Shimane, Japan)). (2014)
- Shoguchi, E. A first assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Joint Aquatic Sciences Meeting 2014. (Oregon Convention Center, USA) (2014)
- Shoguchi, E., Shinzato, C., Hisata, K., and Satoh, N. Decoding the mitochondrial genome in Symbiodinium minutum. In Meeting of Japanese Society of Phycology. (Kyushu University, Fukuoka, Japan). (2015)
- Tagawa, K., Arimoto, A., Satoh, N., and Kawashima, T. A cDNA resource of the hemichordate Ptychodera flava. In The 85th Annual Meeting of the Zoological Society of Japan (Tohoku University, Kawauchi-Kita Campus, Sendai, Miyagi Pref.). (2014)
- Yasuoka, Y. Genomics Study of the Spemann-Mangold Organizer: Occupancy of Tissue-Specific cis-Regulatory Modules by Otx2 and TLE/Groucho for Embryonic Head Specification. 15th International Xenopus Conference. (Pacific Grove, CA, USA) (2014)
- Yasuoka, Y., Koyanagi, R., Shinzato, C., and Satoh, N. Functional analysis of the T-box transcription factor Brachyury in coral gastrula embryos. 47th Annual Meeting of the Japanese Society of Developmental Biologists. (WINC AICHI (Nagoya, Aichi)) (2014)
- Yasuoka, Y., Suzuki, Y., Takahashi, S., Someya, H., Sudou, N., Haramoto, Y., Asashima, M., Sugano, S., and Taira, M. Transcriptional basis of the head organizer in the amphibian gastrula: ChIP seq analysis of the head organizer transcription facotors, Otx2, Lim1 and Gsc. National BioResource Project (NBRP) Symposium - Xenopus tropicalis, Functional genomics research employing Xenopus, The 85th Annual Meeting of the Zoological Society of Japan. (Tohoku University, Kawauchi-Kita Campus, Sendai, Miyagi Pref.) (2014)
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 OIST Marine Genomics Seminar
- Title: Corals under stress - A study of the coral immune system
- Speaker: Dr. Jeroen van de Water (James Cook University)
- Date: 13:00 - 14:00 April 13, 2015
- Venue: OIST Lab1 Level-D Meeting Room (D015), Okinawa, Japan
6.2 OIST Marine Genomics Seminar
- Title: Microbial metagenomics of deepsea brine pools
- Speaker: Dr. Pei-Yuan Qian (Hong Kong University of Science and Technology)
- Date: 15:00 - 16:00 Aug 25, 2014
- Venue: OIST Center Building (C209), Okinawa, Japan
7. Other
Nothing to report.



