FY2012 Annual Report
Marine Genomics Unit
Professor Nori Satoh

Abstract
The genome contains all the genetic information of a given organism. Decoding the genome therefore provides the molecular basis for understanding every biological phenomenon. Since the turn of the 21st century, genomes of various metazoans have been sequenced, and consequently studies progressed efficiently in the fields of evolutionary biology, developmental biology, and environmental biology. The objective of the Marine Genomic Unit is to decode the genomes of target marine invertebrates so as to comprehensively elucidate the molecular mechanisms underlying the (a) environmental responses, (b) development and evolution of marine invertebrates. In 2011, we have decoded genome of the coral Acroporal digitifera and that of the pearl oyster Pinctada fucata, in 2012. With the genomic information, in fiscal year 2012, we have challenged several problems in these fields. The major achievements were as follows.
(a) Environmental genomics:
The coral reefs of the Okinawa islands are amongst the most biologically diverse ecosystems in the world. The key organisms in their establishment, the scleractinian corals, increasingly face a range of human-caused challenges including seawater temperature rises and ocean acidification. To understand better the molecular mechanisms underlying coral biology, we succeeded in deciphering the 420-Mbp-long genome of the coral Acropora digitifera in 2011, and found that the coral innate immunity repertoire is notably more complex than that of the sea anemone. This year, we disclose the extraordinary complexity of the innate immune repertoire of corals, which may in part reflect adaptive evolution to a symbiotic lifestyle in a uniquely complex and challenging environment.
(b) Developmental and evolutionary genomics:
We are interested in the origin and evolution of chordates. We have already analyzed an evolutionary scenario within three groups of chordates. Since chordates originated from common ancestor shared with nonchordate deuterostomes such as echinoderms and hemichordates, a big question remained is an evolutionary link between non-chordate deuterostomes and chordates. This year, we found that the two hemichordates, Saccoglossus kowalevskii and Ptychodera flava both possess 12-Hoxgene clusters, with mir10 between hox4 and hox5, in 550 kb and 452 kb intervals, respectively. Hox clusters of vertebrates and the basal chordate amphioxus have similar organization to the hemichordate cluster, providing genomic evidence for a well-ordered complex in the deuterostome ancestor for the hox1–hox9/10 region.
1. Staff
- Group Leader
- Dr. Takeshi Kawashima
- Dr. Eiichi Shoguchi
- Dr. Chuya Shinzato
- Researcher
- Dr. Mayuko Hamada
- Dr. Ryo Koyanagi
- Dr. Ken Maeda
- Dr. Keisuke Nakashima
- Dr. Sutada Mungpakdee
- Dr. Takeshi Takeuchi
- Dr. Fuki Gyoja
- Dr. Yuuri Yasuoka (JSPS, PD Fellow)
- Technical Staff
- Ms. Kanako Hisata
- Ms. Sakura Kikuchi
- Dr. Makiko Tanaka
- Research Assistant
- Yuki Yasuoka
- Research Administrator
- Ms. Shoko Yamakawa
- Ms. Tomomi Teruya
2. Collaborations
- Theme: Decoding of the coral genomes
- Type of collaboration: Collaboration
- Researchers:
- Dr. David Miller, James Cook University, Australia
- Theme: Chip-sequence analysis of Ci-Bra target genes
- Type of collaboration: Collaboration
- Researchers:
- Dr. Hiroki Takahashi, National Institute for Basic Biology, Japan
- Theme: Analysis of Ciona genes with transgenic lines
- Type of collaboration: Collaboration
- Researchers:
- Dr. Yasunori Sasakura, Shimoda Marine Research Center, University of Tsukuba, Japan
- Theme: Analysis of the evolution of chordate genomes
- Type of collaboration: Collaboration
- Researchers:
- Dr. Asao Fujiyama, National Institute of Genetics, Japan
3. Activities and Findings
3.1 Environmental genomics
To understand better the molecular mechanisms underlying coral biology, we decoded in 2011 the approximately 420-megabase genome of Acropora digitifera using next-generation sequencing technology. This genome contains approximately 23,700 gene models. Innate immunity in corals is of special interest not only in the context of self-defense but also in relation to the establishment and collapse of their obligate symbiosis with dinoflagellates of the genus Symbiodinium.
In innate immunity system of vertebrates, approximately 20 tripartite nucleotide oligomerization domain (NOD)-like receptor proteins that are defined by the presence of a NAIP, CIIA, HET-E and TP1 (NACHT) domain, a C-terminal leucine-rich repeat (LRR) domain, and one of three types of N-terminal effector domain, are known to function as the primary intracellular pattern recognition molecules. Surveying the coral genome revealed not only a larger number of NACHT- and related domain nucleotide-binding adaptor shared by APAF-1, R proteins, and CED-4 (NB-ARC)-encoding loci (~500) than in other metazoans but also surprising diversity of domain combinations among the coral NACHT/NB-ARC-containing proteins; N-terminal effector domains included the apoptosis-related domains caspase recruitment domain (CARD), death effector domain (DED), and Death, and C-terminal repeat domains included LRRs, tetratricopeptide repeats, ankyrin repeats, and WD40 repeats (Figure 1). Many of the predicted coral proteins that contain a NACHT/NB-ARC domain also contain a glycosyl transferase group 1 domain, a novel domain combination first found in metazoans. Phylogenetic analyses suggest that the NACHT/NB-ARC domain inventories of various metazoan lineages, including corals, are largely products of lineage-specific expansions. Many of the NACHT/NB-ARC loci are organized in pairs or triplets in the Acropora genome, suggesting that the large coral NACHT/NB-ARC repertoire has been generated at least in part by tandem duplication. In addition, shuffling of N-terminal effector domains may have occurred after expansions of specific NACHT/ NB-ARC-repeat domain types. These results illustrate the extraordinary complexity of the innate immune repertoire of corals, which may in part reflect adaptive evolution to a symbiotic lifestyle in a uniquely complex and challenging environment.
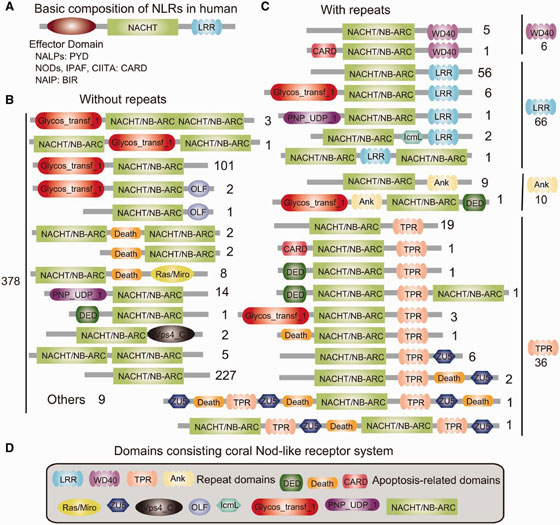
3.2 Developmental and evolutionary genomics
We are interested in the origin and evolution of chordates. Chordates consist of three subphyla, cephalochordates, urochordates, and vertebrates. We have already analyzed an evolutionary scenario within the three groups of chordates, namely, cephalochordates are most basal, and urochordates and vertebrates form a sister group. Since chordates originated from common ancestor shared with nonchordate deuterostomes such as echinoderms and hemichordates, a big question remained is an evolutionary link between non-chordate deuterostomes and chordates. This year, in collaboration with US researchers, challenged this question by comparing Hox cluster genes.
Genomic comparisons of chordates, hemichordates, and echinoderms can inform hypotheses for the evolution of these strikingly different phyla from the last common deuterostome ancestor. Because hox genes play pivotal developmental roles in bilaterian animals, we analyzed the Hox complexes of two hemichordate genomes. We find that Saccoglossus kowalevskii and Ptychodera flava both possess 12-gene clusters, with mir10 between hox4 and hox5, in 550 kb and 452 kb intervals, respectively (Figure 2). Genes hox1–hox9/10 of the clusters are in the same genomic order and transcriptional orientation as their orthologs in chor- dates, with hox1 at the 30 end of the cluster. At the 50 end, each cluster contains three posterior genes specific to Ambulacraria (the hemichordate-echinoderm clade), two forming an inverted terminal pair. In contrast, the echinoderm Strongylocentrotus purpuratus contains a 588 kb cluster of 11 orthologs of the hemichordate genes, ordered differently, plausibly reflecting rearrangements of an ancestral hemichordate-like ambulacrarian cluster (Figure 3). Hox clusters of vertebrates and the basal chordate amphioxus have similar organization to the hemichordate cluster, but with different posterior genes. These results provide genomic evidence for a well-ordered complex in the deuterostome ancestor for the hox1–hox9/10 region, with the number and kind of posterior genes still to be elucidated.
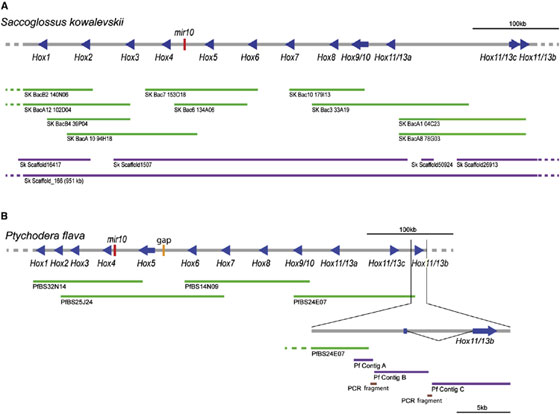
Figure 2. Clustering of Hox Genes in Hemichordate Genomes Genomic regions ∼500 kb-long are shown for (A) Saccoglussus kowalevskii and (B) Ptychodera flava. Both contain the 12 genes Hox1 to Hox11/13b (blue arrows in the direction of transcription). In both, the ten genes Hox1–Hox11/13a are aligned in the same direction, whereas two genes, Hox11/13c and Hox11/13b, are in the opposite direction. Red bars indicate the position of mir10 genes, and the orange bar in (B) indicates a gap. BAC clones are shown as green lines, scaffolds or contigs obtained from whole-genome shotgun reads as purple lines, and PCR-amplified fragments as brown lines.
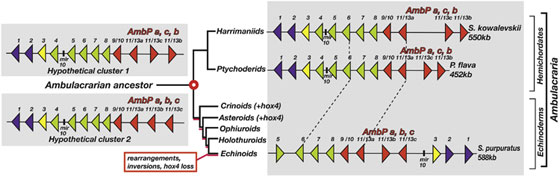
Figure 3. Genomic Organization of Two Hemichordate Hox Clusters and Hypothetical Reconstructions of Hox Cluster of Ambulacrarian Ancestor. Summary of Hox clusters of extant two hemichordates and one echinoderm (Ambulacraria) and two hypothetical reconstructions of the cluster of the ambulacrarian ancestor, arranged in a phylogenetic tree [1]. Hox gene location and transcriptional orientation are indicated by triangles, the colors of which represent the different categories of Hox genes: anterior (purple), group3 (yellow), central (green), and posterior (red). The mir10 locus is indicated by name, between hox4 and hox5. Note the identical organization of the five gene segment (hox6, hox7, hox8, hox9/10, hox11/13a) of the echinoderm (S. purpuratus, sea urchin) and hemichordate clusters (bounded by dashed lines). In the hypothetical cluster 1 (upper left), the minimal ancestral cluster is proposed to be the same as the extant hemichordate clusters, including the inverted hox11/13b,c pair of genes, Then, within the lineage to sea urchins, the cluster underwent rearrangements and gene loss (hox4) as well as an inversion and translocation of hox11/13c. In the hypothetical cluster 2 (lower left), the minimal ancestral cluster is proposed to be like that of hemichordates except that hox11/13b and hox11/13c are in tandem with the other genes, not inverted as a pair. Then within the echinoderm lineage, the cluster underwent rearrangements of hox1–5 and gene loss (hox4), as well as an inversion, but not translocation of hox11/13b, whereas hox11/13b and hox11/13c inverted as a pair in the hemichordate lineage. The proposal of the kinds of hox genes in the ancestral cluster is supported by the hox sequence repertoire across extant Ambulacraria. Whereas hox1–9 (called 9/10 in Ambulacraria) are orthologous to chordate genes 1–9, the ambulacrarian posterior genes 11/13a,b,c differ from vertebrate genes 11–13 and are identified here as AmbPa,b,c to indicate their ambulacrarian-specific ancestry.
4. Publications
4.1 Journals
[2012]
Freeman R, Ikuta T, Wu M, Koyanagi R, Kawashima T, Tagawa K, Humphreys T, Fang GC, Fujiyama A, Saiga H, Lowe C, Worley K, Jenkins J, Schmutz J, Kirschner M, Rokhsar D, Satoh N, Gerhart J. 2012. Identical Genomic Organization of Two Hemichordate Hox Clusters. Current Biology 22: 2053–2058.
Sasakura,Y., Kanda, M., Ikeda, T., Horie, T., Kawai, N., Ogura, Y., Yoshida, Y., Hozumi, A., Satoh, N., and Fujiwara, S. 2012. Retinoic acid-driven Hox1 is required in the epidermis for forming the otic/atrial placode during ascidian metamorphosis. Development 139: 2156-2160.
Satou,Y., SHin-i. T., Kohara, Y., Satoh, N., and Chiba, S. 2012. A genomic overview of short genetic variations in a basal chordate, Ciona intestinalis. BMC Genomics 13:208.
Shinzato, C., Shoguchi, E., Tanaka, M., and Satoh, N. 2012. Fluorescent Protein Candidate Genes in the Coral Acropora digitifera Genome. Zoolog Sci 29: 260-264.
DuBuc, T. Q., Ryan, J. F., Shinzato, C., Satoh, N., and Martindale, M. Q. 2012. Coral comparative genomics reveal expanded Hox Cluster in the Cnidarian–Bilaterian Ancestor. Integrative and Comparative Biology. on line.
Shinzato, C., Hamada, M., Shoguchi, E., Kawashima, T., Satoh, N. 2012. The Repertoire of Chemical Defense Genes in the Coral Acropora digitifera Genome. Zoolog Sci., 29(8):510-7.
Gyoja, F., Kawashima, T. and Satoh, N. 2012. A genomewide survey of bHLH transcription factors in the coral Acropora digitifera identifies three novel orthologous families, pearl, amber, and peridot. Dev Genes Evol 222: 63-76.
Taillebois, L., Maeda, K., Vigne, S., and Keith, P. 2012. Pelagic larval duration of three amphidromous Sicydiinae gobies (Teleostei: Gobioidei) including widespread and endemic species. Ecology of Freshwater Ecology of Freshwater Fish 21: 552-559.
Feutry, P., Tabouret, H., Maeda, K., Pécheyran, C., and Keith, P. 2012. Diadromous life cycle and behavioural plasticity in freshwater and estuarine Kuhliidae species (Teleostei) revealed by otolith microchemistry. Aquatic Biology, 15: 195-204.
Kondo, M., Maeda, K., Yamasaki, N., and Tachihara, K. 2012. Spawning habitat and early development of Luciogobius ryukyuensis (Gobiidae). Enviromental biology of Fishes 95(2): 291-300.
Sato, A., Satoh, N. and Bishop, J. D. 2012. Field identification of ‘types’ A and B of the ascidian Ciona intestinalis in a region of sympatry. Marine Biology 159: 1611-1619.
Morino Y, Koga H, Tachibana K, Shoguchi E, Kiyomoto M, Wada H. 2012. Heterochronic activation of VEGF signaling and the evolution of the skeleton in echinoderm pluteus larvae. Evolution and Development 14(5):428-36.
Kume S, Ueki T, Matsuoka H, Hamada M, Satoh N, Michibata H. 2012. Differential gene regulation by V(IV) and V (V) ions in the branchial sac, intestine, and blood cells of a vanadium-rich ascidian, Ciona intestinalis. Biometals Oct;25(5):1037-50.
[2013]
Hamada, M., Shoguchi, E., Shinzato, C., Kawashima, T., Miller, D. J., Satoh, N. 2012. The complex NOD-like receptor repertoire of the coral Acropora digitifera includes novel domain combinations. Mol Biol Evol. 30(1):167-76.
Shoguchi, E., Tanaka, M., Shinzato, C., Kawashima, T., Satoh, N. 2012. A genome-wide survey of photoreceptor and circadian genes in the coral, Acropora digitifera. Gene 515: 426–431.
4.2 Books and other one-time publications
Satoh, N. 2013 Ciona: a Model for Developmental Genomics. Encyclopedia of Life Sciences.
Takeuchi, T., Kawashima, T., Koyanagi, R., Satoh, N. 2013. The genome of the Japanese pearl oyster, Pinctada fucata. In "Recent Advances in Pearl Research".Eds. S. Watabe, K. Maeyama, H. Nagasawa, TERRAPUB, Tokyo
4.3 Oral and Poster Presentations
Hamada M, Kürn U, Schröder K, Fraune S, Satoh N, and Bosch TCG. 2013. The Hydra viridissima /Chlorella symbiosis revisited:Understanding a complex team of players at the base of animal evolution. In International Joint Meeting of the German Society for Cell Biology (DGZ) and the German Society for Developmental Biology (GfE). (Heidelberg, Germany).
Satoh N. 2012. Decoding Genomes of Marine Organisms for Basic and Applied Marine Biosciences. In BIT's 2nd Annual World Congress of Marine Biotechnology 2012. (World Expo Center, Dalian, China).
Satoh N. 2012. Evolutionary Genomics. In NGS4MG: Next Generation Sequencing for Marine Genomics Symposium. (Vigo, Spain).
Satoh N. 2012. Developmental mechanisms underlying the origin of chordates. In Asia-Pacific Developmental Biology Conference (APDBC 2012). (Taipei, Taiwan).
Yasuoka Y, Suzuki Y, Takahashi S, Sudou N, Haramoto Y, Cho KW, et al. 2012. Head specification mediated by Otx2 partner with Lim1 or Gsc and control expression of thousands of target genes in a massively parallel fashion. In 14th Xenopus International Conference. (Giens Peninsula, France).
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 OIST Marine Genomics Seminar
- Title: New vectors for gene delivery: Human and mouse artificial chromosomes.
- Speaker: Dr. Mitsuo Oshimura (Tottori University, Japan)
- Date: 18:10 – 18:40, Feb 15, 2013
- Venue: OIST Lab1 Level-D Meeting Room 1 (D015), Okinawa, Japan
6.2 OIST Marine Genomics Seminar
- Title: Toward safe and effective gene- and cell-therapies using human artificial chromosomes and stem cells.
- Speaker: Dr. Narumi Uno (Tottori University, Japan)
- Date: 18:40 – 19:10 Feb 15, 2013
- Venue: OIST Lab1 Level-D Meeting Room 1 (D015), Okinawa, Japan
6.3 OIST Winter Course "Evolution of Complex Systems" (OWECS) 2012
- Date: 6-11 December, 2010
- Venue: OIST Seaside House, Okinawa, Japan
- Co-organizers
- Dr. Sydney Brenner, OIST, Japan
- Dr. Michael Levine, University of California, Berkeley, USA
- Speakers:
- Michael Levine (University of California, Berkeley, USA)
- Robb Krumlauf (Stowers Institute for Medical Research, USA)
- Nipam H. Patel (University of California, Berkeley, USA)
- Daniel Rokhsar (University of California, Berkeley, USA)
- Nori Satoh (OIST, Japan)
- J. K. Sky Yu (Academia Sinica, Taiwan)
- John Gerhart (University of California, Berkeley, USA)
- Yi-Hsien Su (Academia Sinica, Taiwan)
- Hiroshi Wada (University of Tsukuba, Japan)
- Shigehiro Kuraku (Riken CDB, Japan)
- Robert Sinclair (OIST, Japan)
- Chris Amemiya (Benaroya Research Institute at Virginia Mason, USA)
7. Other
Nothing to report.



