FY2021 Annual Report
FY2021 Annual Report
Abstract
The genome contains all the genetic information of a given organism. Decoding the genome provides the molecular basis for understanding every biological phenomenon. Since 2009, the Marine Genomics Unit (MGU) has conducted research in the realm of genome-based biological sciences. By sequencing genomes of target marine organisms (mainly invertebrates), we wish to understand genetic and developmental mechanisms underlying evolution and diversification of marine organisms. The major research fields are (a) evolutionary and developmental genomics of marine invertebrates, (b) environmental genomics of coral reefs, and (c) functional genomics of marine organisms including pearl oyster and algae. To date, we have reported draft genomes of a coral in 2011, a pearl oyster in 2012, and coral-symbiotic dinoflagellate (Breviolum) in 2013. We further decoded genomes of hemichordates and a brachiopod in 2015; a brown alga (Okinawa-mozuku) in 2016; Crown-Thorns-Starfish in 2017; a nemertean, phoronid, and two dinoflagellate clades in 2018; jellyfish, dicyemid, acoel flatworm, siphonous macroalga (umi-budo), brown alga (ito-mozuku) in 2019; and hydra and four four strains of “Okinawa mozuku” brown alga in 2020. This year we have reported sequenced genome of 19 coral species (collaboration with Tokyo Univ.), the kuruma shrinp (collaboration with Tokyo U. of Marine Sci. Tech.), and nearly complete genome of the tunicate Ciona (collaboration with Kyoto U.). In addition, we have advanced genome-based coral research this year, and one research result shall be reported below.

1. Staffs and Students
- Professor Noriyuki Satoh
- Staff Scientists
- Eiichi Shoguchi (Group Leader)
- Keisuke Nakashima
- Konstantin Khalturin
- Takeshi Takeuchi
- Koki Nishitsuji
- Yuna Zayasu
- Postdoctoral Scholars
- Hitoshi Tominaga
- Technical Staffs
- Kanako Hisata
- Sakura Kikuchi
- Haruhi Narisoko
- Research Assistants
- Yoshie Nishitsuji (AIMS project)
- Mayuki Suwa (Part time)
- Research Administrators
- Shoko Yamakawa
- Tomomi Teruya
- Students
- Ph.D students (co-superviser)
- Kun-Lung Li (Supervisor: Assistant Prof. Watanabe, H.)
- Rio Kashimoto (Supervisor: Prof. Laudet, V.)
- Ph.D students (co-superviser)
2. Collaborations
2-1 Genome scientific studies of chordate evolution
- Type of collaboration: Scientific collaboration
- Researchers: Prof. Daniel Rokshar, OIST
2-2 Genome sequencing of marine invertebrates at haplotype-resolution level
- Type of collaboration: Scientific collaboration
- Researchers: Prof. Gene Myers, OIST
2-3. Molecular biological study of COTS communications
- Type of collaboration: Scientific collaboration
- Researchers: Prof. Scott Cummins, Univ. Sunshine Coast, Australia
2-4. Genome scientific study of dinoflagellates
- Type of collaboration: Scientific collaboration
- Researchers: Prof. Pengchen Fu, Hainan University
2-5. Genome scientific study of coral-dinoflagellate symbiosis
- Type of collaboration: Scientific collaboration
- Researchers: Profs. Shigeki Fujiwara & Kaz Kawamura, Kochi University
2-6. Genome scientific study of left-right asymmetry of snails
- Type of collaboration: Scientific collaboration
- Researchers: Prof. Takehiro Asami, Shinshu University
2-7. Genome scientific study of acoel development
- Type of collaboration: Scientific collaboration
- Researchers: Profs. Kunifumi Tagawa & Tatsuya Ueki, Hiroshima University
3. Research activities and findings
We report here an interesting result of research collaboration with local peoples involved in coral reef preservation of the Okinawa coast. That is, color polymorphism of the coral, Acropora tenuis, is likely involved in different responses to environmental stress via different expression profiles of fluorescent-protein genes (see publication #4).
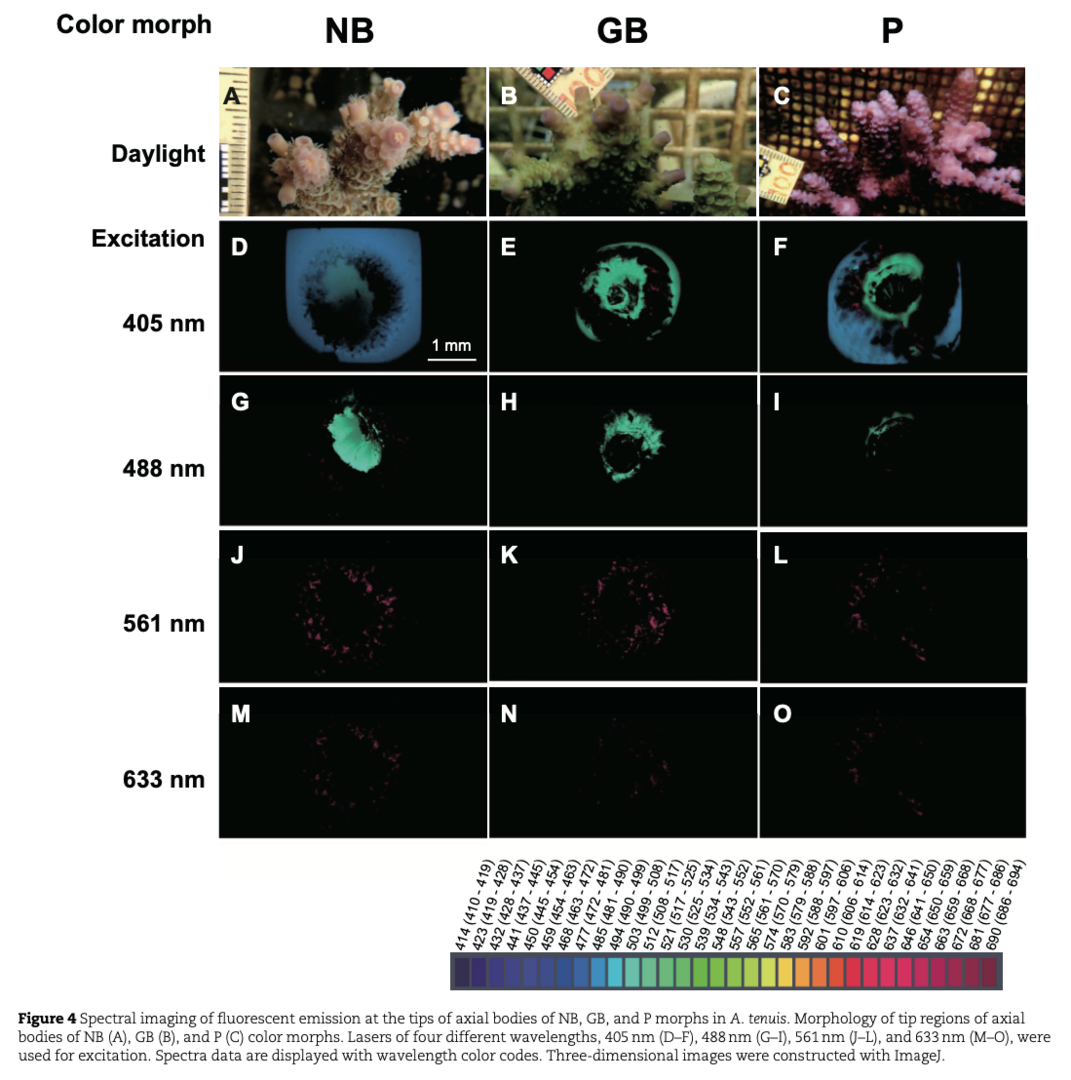
Corals of the family Acroporidae are key structural components of reefs that support the most diverse marine ecosystems. Due to increasing anthropogenic stresses, coral reefs are in decline. Along the coast of Okinawa, Japan, three different color morphs of Acropora tenuis have been recognized for decades. These include brown (N morph), yellow green (G), and purple (P) forms (Fig. 1). The tips of axial polyps of each morph exhibit specific fluorescence spectra (Fig. 4). This attribute is inherited asexually, and color morphs do not change seasonally.

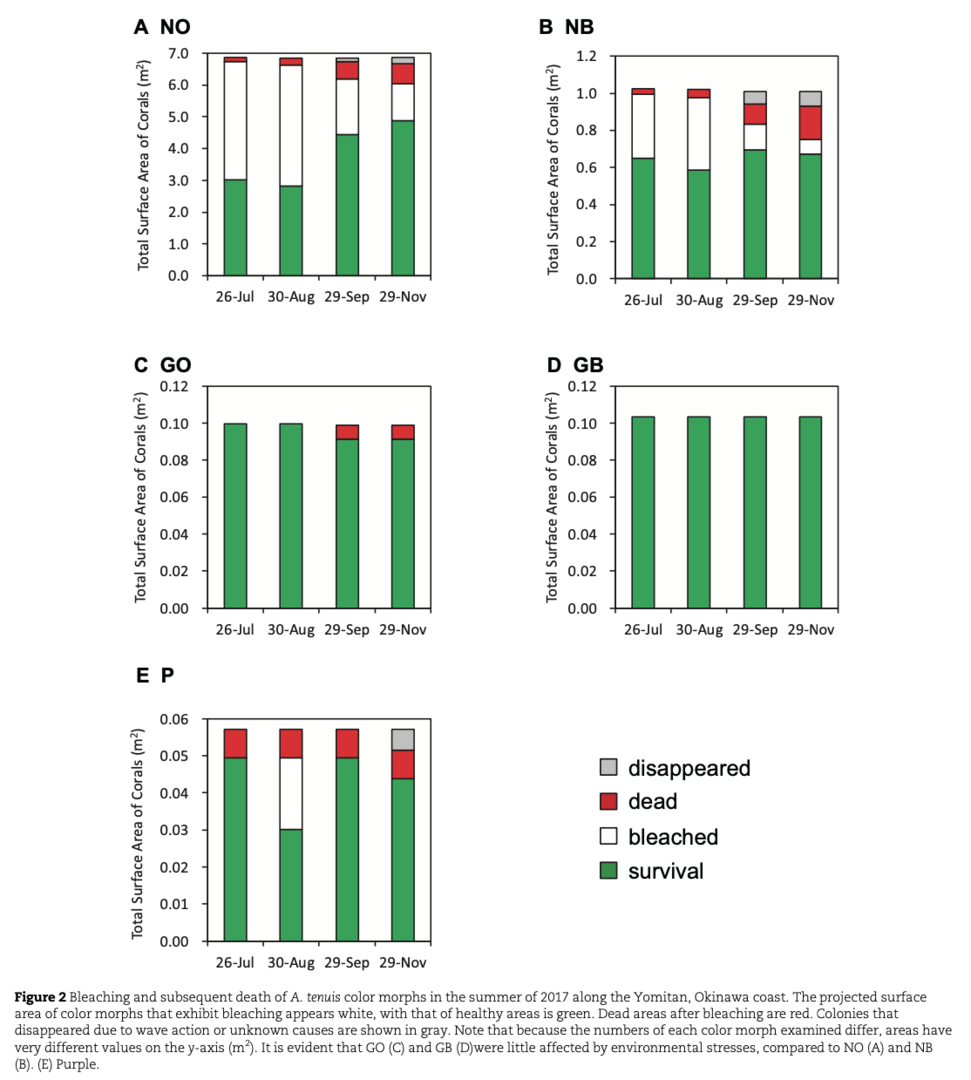
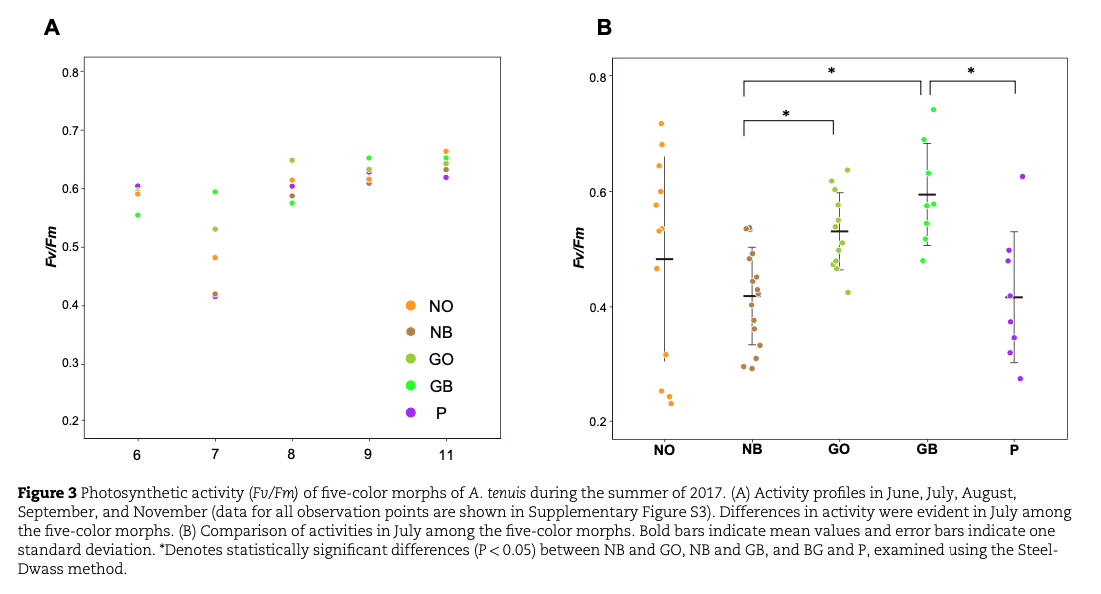
In Okinawa Prefecture, during the summer of 2017, N and P morphs experienced bleaching, in which many N morphs died. Dinoflagellates (Symbiodiniaceae) are essential partners of scleractinian corals, and photosynthetic activity of symbionts was reduced in N and P morphs. In contrast, G morphs successfully withstood the stress (Figs. 2 and 3).
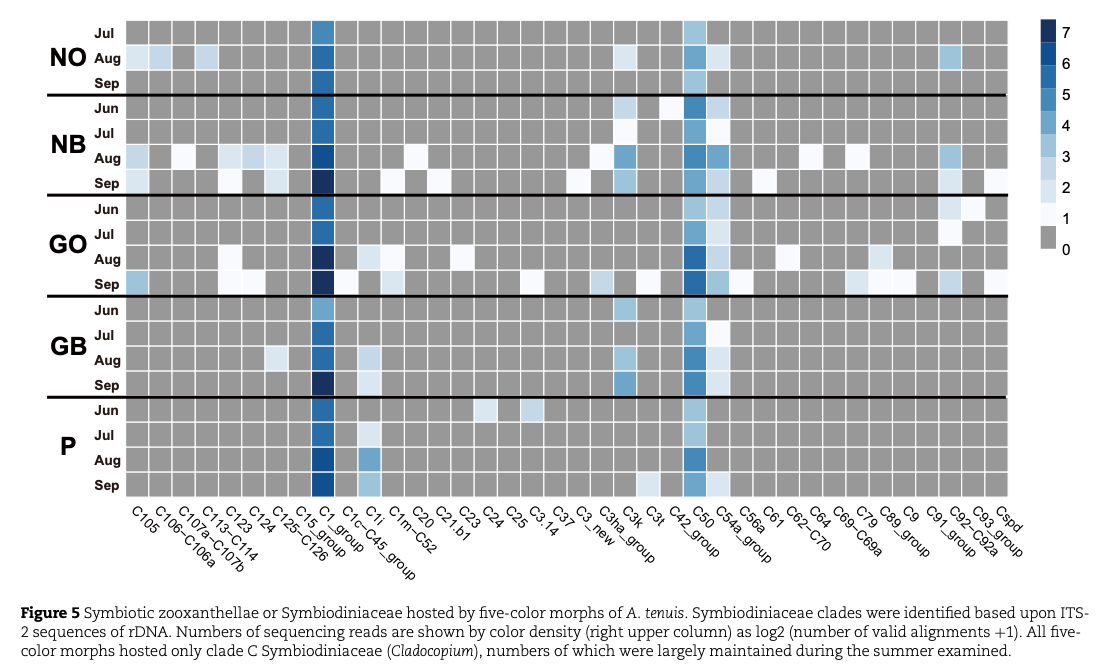
Examination of the clade and type of Symbiodiniaceae indicated that the three color-morphs host similar sets of Clade-C symbionts, suggesting that beaching of N and P morphs is unlikely attributable to differences in the clade of Symbiodiniaceae the color morphs hosted (Fig. 5).
Fluorescent proteins play pivotal roles in physiological regulation of corals. Since the A. tenuis genome has been decoded, we identified five genes for green fluorescent proteins (GFPs), two for cyan fluorescent proteins (CFPs), three for red fluorescent proteins (RFPs), and seven genes for chromoprotein (ChrP) (Fig.6).
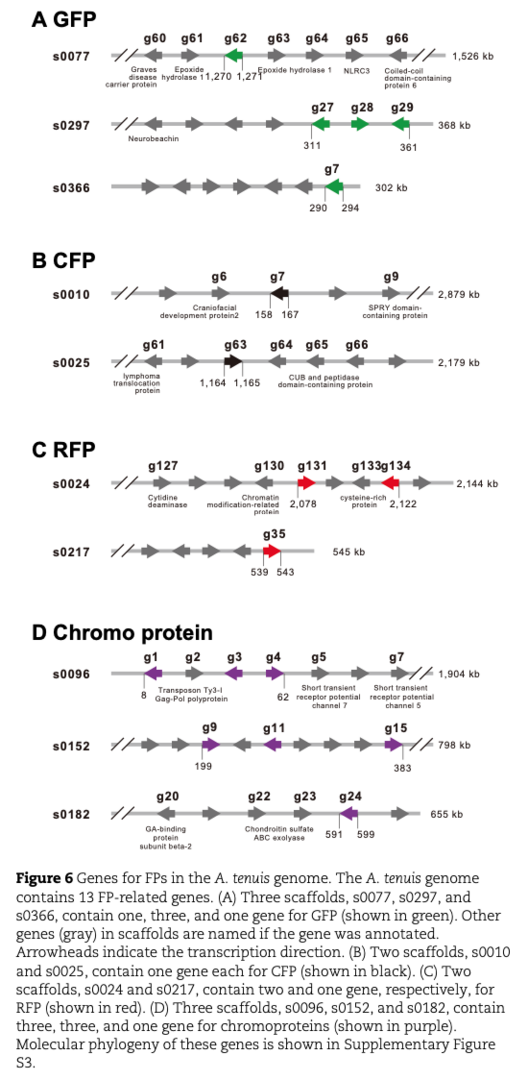
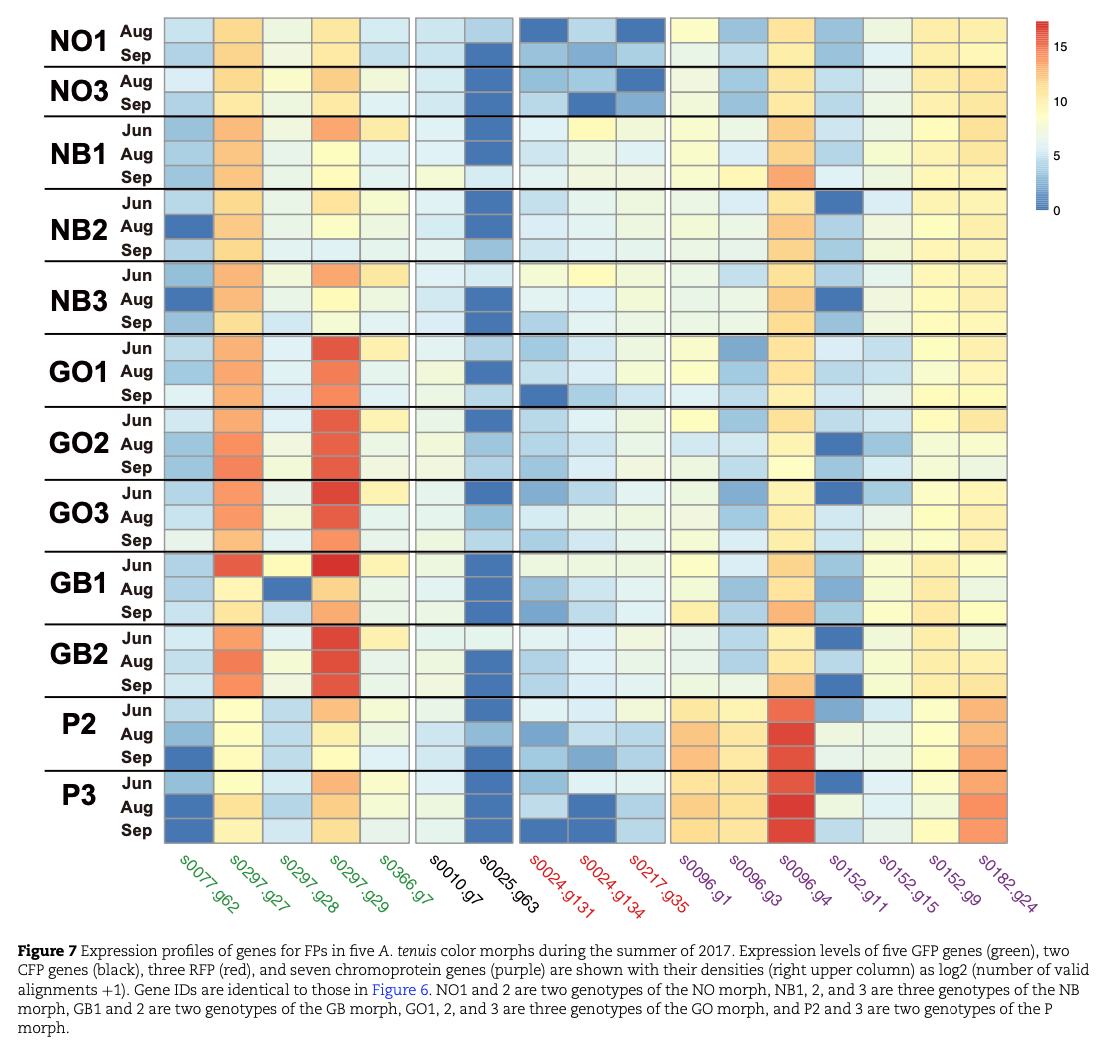
A summer survey of gene expression profiles under outdoor aquarium conditions demonstrated that (a) expression of CFP and REP was quite low during the summer in all three morphs, (b) P morphs expressed higher levels of ChrP than N and G morphs, (c) both N and G morphs expressed GFP more highly than P morphs, and (d) GFP expression in N morphs was reduced during summer whereas G morphs maintained high levels of GFP expression throughout the summer (Fig. 7).
Although further studies are required to understand the biological significance of these color morphs of A. tenuis, our results suggest that thermal stress resistance is modified by genetic mechanisms that coincidentally lead to diversification of color morphs of this coral.
4. Publications
(a) Environmental Genomics
- Shinzato, C., Takeuchi, T., Yoshioka, Y., Tada, I., Kanda, M., Broussard, C., Iguchi, A., Kusakabe, M., Marin, F., Satoh, N., Inoue, M.
Whole-genome sequencing highlights conservative genomic strategies of a stress-tolerant, long-lived scleractinian coral, Porites australiensis Vaughan, 1918. Genome Biology and Evolution,13(12):evab270. doi: 10.1093/gbe/evab270. (2021). - Kawamura, K., Sekida, S., Nishitsuji, K., Shoguchi, E., Hisata, K., Fujiwara, S., Satoh, N.
In vitro symbiosis of reef-building coral cells with photosynthetic dinoflagellates.
Frontiers in Marine Science, doi:10.3389/fmars.2021.706308 (2021). - Shinzato, C., Khalturin, K., Inoue, J., Zayasu, Y., Kanda, M., Kawamitsu, M., Yoshioka, Y., Yamashita, H., Suzuki, G., Satoh, N.
Eighteen coral genomes reveal the evolutionary origin of Acropora strategies to accommodate environmental changes.
Molecular Biology and Evolution, 38: 16-30. (2021). - Satoh, N., Kinjo, K., Kohei Shintaku, K., Kezuka, D., Ishimori, H., Yokokura, A., Hagiwara, K., Hisata, K., Kawamitsu, M., Koizumi, K., Shinzato, C., Zayasu, Y.
Color morphs of the coral, Acropora tenuis, show different responses to environmental stress and different expression profiles of fluorescent-protein genes.
G3: Genes|Genomes|Genetics, 11, jkab018 (2021). - Shinzato, C., Narisoko, H., Nishitsuji, N., Nagata, T., Satoh, N., Inoue, J.
Novel mitochondrial DNA markers for scleractinian corals and generic-level environmental DNA metabarcoding.
Frontier in Marine Science 8:758207. doi: 10.3389/fmars.2021.758207 (2021). - Kawamura, K., Nishitsuji, K., Shoguchi, E., Fujiwara, S., Satoh, N.
Establishing sustainable cell lines of a coral, Acropora tenuis.
Marine Biotechnology, doi: 10.1007/s10126-021-10031-w (2021). - Mason, B., Cooke, I., Moya, A., Augustin, R., Lin, M., Satoh, N., Bosch, T., Bourne, D., Hayward, D., Andrade, N., Forêt, S., Ying, H., Ball, E., Miller, D.J.
AmAMP1 from Acropora millepora and damicornin define a family of coral-specific antimicrobial peptides related to the Shk toxins of sea anemones.
Developmental and Comparative Immunology, 114: 103866 (2021). - Zayasu, Y., Takeuchi, T., Nagata, T., Kanai, M., Fujie, M., Kawamitsu, M. Chinen, W. Shinzato, C., Satoh, N.
Genome-wide SNP genotyping reveals hidden population structure of an acroporid species at a subtropical coral island: Implications for coral restoration.
Aquatic Conservation: Marine and Freshwater Ecosystems, 1–11 (2021). - Sassa, M., Takagi, T., Kinjo, A., Yoshioka, Y., Zayasu, Y., Shinzato, C., Kanda, S., Murakami-Sugihara, N., Shirai, K., Inoue, K.
Divalent metal transporter-related protein restricts animals to marine habitats.
Communication Biology 4, 463 (2021).
(b) Developmental and Evolutionary Genomics
- Satoh, N., Tominaga, H., Kiyomoto, M., Hisata, K., Inoue, J., Nishitsuji, K.
A preliminary single-cell RNA-seq analysis of embryonic cells that express Brachyury in the amphioxus, Branchiostoma japonicum.
Frontiers in Cell Developmental Biology doi:10.3389/fcell.2021.696875 (2021). - Kashimoto, R., Hisata, K., Shinzato, C., Satoh, N., Shoguchi, E.
Expansion and diversification of fluorescent protein genes in fifteen Acropora species during evolution of acroporid corals.
Genes, 12: 397 (2021). - Satou, Y., Sato, A., Yasuo, H., Mihirogi, Y., Bishop, J., Fujie, M., Kawamitsu, M., Hisata, K., Satoh, N.
Chromosomal inversion polymorphisms in two sympatric ascidian lineages.
Genome Biology and Evolution, evab068 (2021).
(c) Functional Genomics
- Takeuchi, T., Fujie, M., Koyanagi, R., Plasseraud, L., Ziegler-Devin, I., Brosse, N., Broussard, C., Satoh, N., Marin, F.
The ‘shellome’ of the crocus clam Tridacna crocea emphasizes essential components of mollusk shell biomineralization.
Frontiers in Genetics, doi:10.3389/fgene.2021.674539 (2021). - Shoguchi, E.
Gene clusters for biosynthesis of mycosporine-like amino acids in dinoflagellate nuclear genomes: Possible recent horizontal gene transfer between species of Symbiodiniaceae (Dinophyceae).
Journal of Phycology. (2021) - Kawato, S., Nishitsuji, K., Arimoto, A., Hisata, K., Kawamitsu, M., Nozaki, R., Kondo, H., Shinzato, C., Ohira, T., Satoh, N., Shoguchi, E., Hirono, I.
Genome and transcriptome assemblies of the kuruma shrimp, Marsupenaeus japonicus.
G3: Genes|Genomes|Genetics 11:jkab268 (2021).
(d) Sequencing collaboration
- Maeda, T., Takahashi, S., Yoshida, T., Shimamura, S., Takaki, Y., Nagai, Y., Toyoda, A., Suzuki, Y., Arimoto, A., Ishii, H., Satoh, N., Nishiyama, T., Hasebe, M., Maruyama, T., Minagawa, J., Obokata, J., Shigenobu, S.
Chloroplast acquisition without the gene transfer in kleptoplastic sea slugs, Plakobranchus ocellatus.
eLife, 10:e60176. doi: 10.7554/eLife.60176 (2021). - Patwary, Z.P., Paul, N.A., Nishitsuji, K., Campbell, A.H., Shoguchi, E., Zhao, M., Cummins, S.F.
Application of omics research in seaweeds with a focus on red seaweeds.
Brief in Functional Genomics, elab023. doi: 10.1093/bfgp/elab023 (2021). - Okubo, S., Ena, E., Okuda, A., Kozone, I., Hashimoto, J., Nishitsuji, Y., Fujie, M., Satoh, N., Ikeda, H., Shin-Ya, K.
Identification of functional cytochrome P450 and ferredoxin from Streptomyces sp. EAS-AB2608 by transcriptional analysis and their heterologous expression.
Applied Microbiology and Biotechnology, 105: 4177-4187. doi: 10.1007/s00253-021-11304-z. (2021) - Yoneda, Y., Yamamoto, K., Makino, A., Tanaka, Y., Meng, X.Y., Hashimoto, J., Shin-ya, K., Satoh, N., Fujie, M., Toyama, T., Mori, K., Ike, M., Morikawa, M., Kamagata, Y., Tamaki, H.
Novel plant-associated Acidobacteria promotes growth of common floating aquatic plants, duckweeds.
Microorganisms 9:1133. doi.org/10.3390/microorganisms9061133 (2021). - Maeda, K., Shinzato, C., Koyanagi, R., Kunishima, T., Kobayashi, H., Satoh, N., Palla, H.P.
Two new species of Rhinogobius (Gobiiformes: Oxudercidae) from Palawan, Philippines, with their phylogenetic placement. Zootaxa, 5068, 81–98 (2021).
5. Seminar
No seminars in FY2021 due to COVID-2019.



