FY2020 Annual Report
Abstract
The genome contains all the genetic information of a given organism. Decoding the genome therefore provides the molecular basis for understanding every biological phenomenon. Since 2009, the Marine Genomics Unit (MGU) has conducted research in the realm of genome-based biological sciences. By decoding genomes of target marine organisms (mainly invertebrates), we wish to understand genetic and developmental mechanisms underlying diversity of marine organisms. The major research fields are (a) evolutionary and developmental genomics of marine invertebrates, (b) environmental genomics of coral reefs, and (c) functional genomics of marine organisms, including pearl oyster and algae. To date, we have decoded genome of a coral in 2011, a pearl oyster in 2012, and symbiotic dinoflagellate (Breviolum) in 2013. We further decoded genomes of hemichordates and a brachiopod in 2015, a brown alga (Okinawa-mozuku) in 2016, Crown-Thorns-Starfish in 2017, a nemertean, phoronid, and two dinoflagellate clades in 2018, and jellyfish, dicyemid, acoel flatworm, siphonous macroalga (umi-budo), and brown alga (ito-mozuku) in 2019. This year we obtained genome sequences of a hydra, a Durusdinium (coral symbiotic dinoflagellate), and four strains of “Okinawa mozuku” brown alga. A main result of this year research shall be reported below.
1. Staffs and Students
- Professor Noriyuki Satoh
- Staff Scientists
- Eiichi Shoguchi (Group Leader)
- Keisuke Nakashima
- Konstantin Khalturin
- Takeshi Takeuchi
- Koki Nishitsuji
- Yuna Zayasu
- Postdoctoral Scholars
- Hitoshi Tominaga
- Technical Staffs
- Kanako Hisata
- Sakura Kikuchi
- Haruhi Narisoko
- Research Assistants
- Yoshie Nishitsuji (AIMS project)
- Mayuki Suwa (Part time)
- Research Administrators
- Shoko Yamakawa
- Tomomi Teruya
- Students
- Ph.D students (co-superviser)
- Kun-Lung Li (Supervisor: Assistant Prof. Watanabe, H.)
- Christina Ripken (Supervisor: Prof. Shannon, N.)
- Lab rotation students
- Margarita Dronova
- Michael Izumiyama
- Internship Students
- Nanako Okabe (ICU)
- Ph.D students (co-superviser)
2. Collaborations
2-1 Genome scientific studies of chordate evolution
- Type of collaboration: Scientific collaboration
- Researchers: Prof. Daniel Rokshar, OIST
2-2. Molecular biological study of COTS communications
- Type of collaboration: Scientific collaboration
- Researchers: Prof. Scott Cummins, Univ. Sunshine Coast, Australia
2-3. Genome scientific study of dinoflagellates
- Type of collaboration: Scientific collaboration
- Researchers: Prof. Pengchen Fu, Hainan University
2-4. Genome scientific study of coral-dinoflagellate symbiosis
- Type of collaboration: Scientific collaboration
- Researchers: Profs. Shigeki Fujiwara & Kaz Kawamura, Kochi University
2-5. Genome scientific study of left-right asymmetry of snails
- Type of collaboration: Scientific collaboration
- Researchers: Prof. Takehiro Asami, Shinshu University
2-6. Genome scientific study of acoel development
- Type of collaboration: Scientific collaboration
- Researchers: Profs. Kunifumi Tagawa & Tatsuya Ueki, Hiroshima University
3. Research activities and findings
We report here decoding genomes of four strains of “Okinawa mozuku” brown alga in 2020. The brown alga, Cladosiphon okamuranus, is one of the most important edible seaweeds, and it is cultivated for market primarily in Okinawa, Japan. Four strains, denominated the S-strain, the K-strain, the O-strain and the C-strain, with distinctively different morphologies, have been cultivated commercially since the early 2000s (Fig. 1). We previously reported a draft genome of the S-strain. To facilitate studies of seaweed biology for future aquaculture, we here decoded and analyzed genomes of the other three strains (K, O, and C).
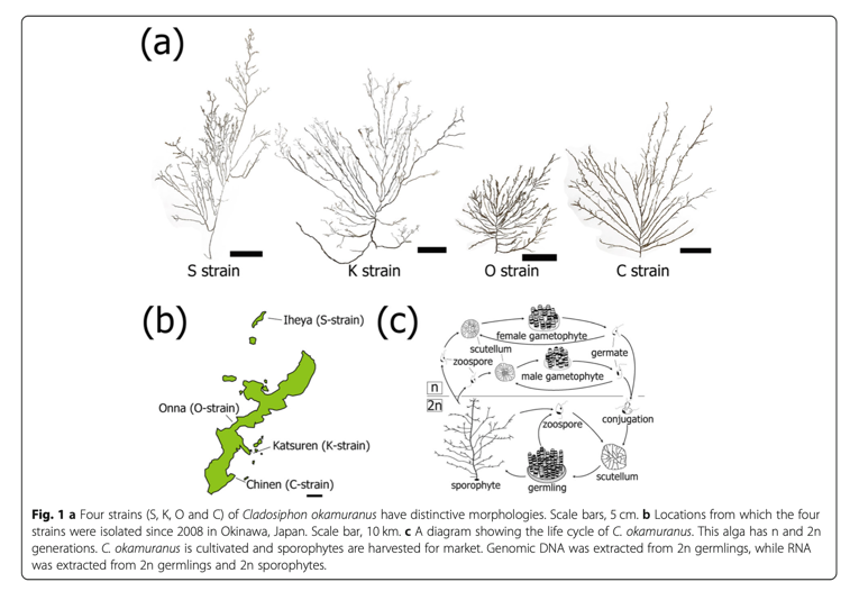
The genome size and the estimated number of genes in the S-strain (ver. 2) were 130 Mbp and 12,999 genes, the K-strain, 135 Mbp and 12,511 genes, the O-strain, 140 Mbp and 12,548 genes, and the C-strain, 143 Mbp and 12,182 genes, respecitively. Molecular phylogenies, using mitochondrial and nuclear genes, showed that the S-strain diverged first, followed by the K- strain, and most recently the C- and O-strains.
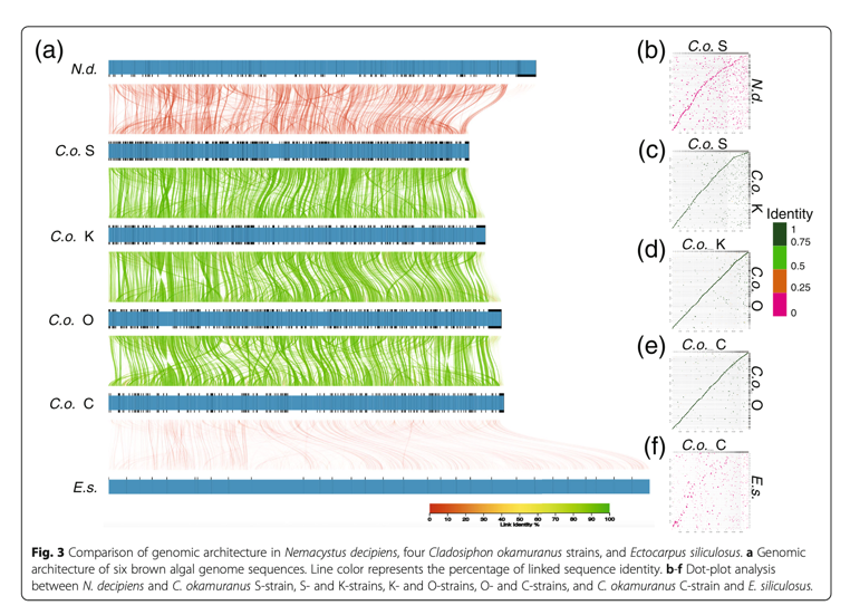
Comparisons of genome architecture among the four strains document the frequent occurrence of inversions (Fig. 3). In addition to gene acquisitions and losses, the S-, K-, O-, and C- strains possess 457, 344, 367, and 262 gene families unique to each strain, respectively. Comprehensive Blast searches showed that most genes have no sequence similarity to any entries in the non-redundant protein sequence database, although GO annotation suggested that they likely function in relation to molecular and biological processes and cellular components.
Due to global environmental changes, including temperature increases, acidification, and pollution, brown algal aquaculture is facing critical challenges. Genomic and phylogenetic information reported by the present research provides useful tools for isolation of novel strains.
4. Publications
(a) Developmental and Evolutionary Genomics
- Hozumi, A., Matsunobu, S., Mita, K., Treen, N., Sugihara, T., Horie, T., Sakuma, T., Yamamoto, T., Shiraishi, A., Hamada, M., Satoh, N., Sakurai, K., Satake, Honoo, Sasakura, Y.
GABA-induced GnRH release triggers chordate metamorphosis
Current Biology, 30:1555-1561 (2020) - Onuma, T.A., Hayashi, M., Gyoja, F., Kishi, K., Wang, K., Nishida, H.
A chordate species lacking Nodal utilizes calcium oscillation and Bmp for left-right patterning.
Proc Natl Acad Sci U S A., 117(8):4188-4198 (2020) - Ilsley, G.R., Suyama, R., Noda, T., Satoh, N., Luscombe, N.M.
Finding cell-specific expression patterns in the early Ciona embryo with single-cell RNA-seq
Scientific Reports, 10:4961 (2020) - Simakov, O., Marlétaz, F., Yue, J.X., O'Connell, B., Jenkins, J., Brandt, A., Calef, R., Tung, C.H., Huang, T.K., Schmutz, J., Satoh, N., Yu, J.K., Putnam, N.H., Green, R.E., Rokhsar, D.S.
Deeply conserved syntheny resolves early events in vertebrate evolution
Nature Ecology & Evolution, 4(6): 820–830 (2020) - Yasuoka, Y.
Morphogenetic mechanisms forming the notochord rod: The turgor pressure‐sheath strength model.
Develop Growth Differ. 62(6):379-390. DOI: 10.1111/dgd.12665 - Li, K.L., Nakashima, K., Inoue, J., Satoh, N.
Phylogenetic analyses of Glycosyl Hydrolase Family 6 genes in tunicates: Possible horizontal transfer
Genes 11, 937, doi:10.3390/genes11080937 - Hamada, M., Satoh, N., Khalturin, K.
A Reference Genome from the symbiotic hydrozoan, Hydra viridissima
G3: Genes|Genomes|Genetics, 10(11):3883-3895. (2020) - Nesterenko, M.A., Starunov, V.V., Shchenkov, S.V., Maslova, A.R., Denisova, S.A., Granovich, A.I., Dobrovolskij, A.A., Khalturin, K.
Molecular signatures of the rediae, cercariae and adult stages in the complex life cycles of parasitic flatworms (Digenea: Psilostomatidae)
Parasites & Vectors volume 13, Article number: 559 (2020) - Zhao, R., Takeuchi, T., Koyanagi, R., Villar-Briones, A., Yamada, L., Sawada, H., Ishikawa, A., Iwanaga, S., Nagai, K., Che, Y., Satoh, N., Endo, K.
Phylogenetic comparisons reveal mosaic histories of larval and adult shell matrix protein deployment in pteriomorph bivalves.
Sci Rep 10, 22140 (2020).
(b) Environmental Genomics
- Shoguchi, E., Yoshioka, Y., Shinzato, C., Arimoto, A., Bhattacharya, D., Satoh, N.
Correlation between organelle genetic variation and RNA editing in dinoflagellates associated with the coral Acropora digitifera
Genome Biology and Evolution, 12:203-209 (2020) - Inoue, J., Hisata, K., Yasuda, N., Satoh, N.
An investigation into the genetic history of Japanese populations of three starfish, Acanthaster planci, Linckia laevigata, and Asterias amurensis, based on complete mitochondrial DNA sequences
G3: Genes|Genomes|Genetics, 10: 2519-2528 (2020) - Takagi, T., Yoshioka, Y., Zayasu, Y., Satoh, N., Shinzato, C.
Transcriptome analyses of immune system behaviors in primary polyp of coral Acropora digitifera exposed to the bacterial pathogen Vibrio coralliilyticus under thermal loading
Marine Biotechnology, 22:748-759. (2020) - Selmoni, O., Lecellier, G., Ainley, L., Collin, A., Doucet, R., Dubousquet, V., Feremaito, H., Waia, E.I., Kininmonth, S.J., Magalon, H., Malimali, S., Maugateau, A., Meibom, A., Mosese, S., Rene-Trouillefou, M., Satoh, N., van Oppen, M.J.H., Xozame, A., Yekawene, M., Joost, S., Berteaux-Lecellier, V.
Using modern conservation tools for innovative management of coral reefs: the MANACO Consortium Journal: Frontiers in Marine Science, section Marine Conservation and Sustainability
Frontiers in Marine Science, 7:609 - Shoguchi, E., Beedessee, G., Hisata, K., Tada, I., Narisoko, H., Satoh, N., Kawachi, M., Shinzato, C.
A New Dinoflagellate Genome Illuminates a Conserved Gene Cluster Involved in Sunscreen Biosynthesis
Genome Biology and Evolution, 13:evaa235. - Yoshioka, Y., Yamashita, H., Suzuku, G., Zayasu, Y., Tada I., Kanda, M., Satoh, N., Shoguchi, E., Shinzato, C.
Whole-genome transcriptome analyses of native symbionts reveal host coral genomic novelties for establishing coral-algae symbioses.
Genome Biology and Evolution, 13:evaa240.
(c) Functional Genomics
- Takeuchi, T., Masaoka, T., Aoki, H., Koyanagi, R., Fujie, M., Satoh, N.
Divergent northern and southern populations and demographic history of the pearl oyster in the western Pacific revealed with genomic SNPs
Evolutionary Applications, 13: 838–854 (2020) - Choi, J.W., Graf, L., Peters, A.F., Cock, J.M., Nishitsuji, K., Arimoto, A., Shoguchi, E., Nagasato, C., Choi, C.G., Yoon, H.S.
Organelle inheritance and genome architecture variation in isogamous brown algae.
Sci Rep. 10:2048. doi: 10.1038/s41598-020-58817-7 - Ishikawa, A., Shimizu, K., Isowa, Y., Takeuchi, T., Zhao, R., Kito, K., Fujie, M., Satoh, N., Endo, K.
Functional shell matrix proteins tentatively identified by asymmetric snail shell morphology.
Scientific Reports, 10:9768. - Nishitsuji, K., Arimoto, A., Yonashiro, Y., Hisata, K., Fujie, M., Kawamitsu, M., Shoguchi, E., Satoh, N.
Comparative genomics of four strains of the edible brown alga, Cladosiphon okamuranus
BMC Genomics, 21: 422 (2020) - Hashimoto, T., Kozone, I., Hashimoto, J., Suenaga, H., Fujie, M., Satoh, N., Ikeda, H., Shin-Ya, K.
Identification, Cloning and Heterologous Expression of Biosynthetic Gene Cluster for Desertomycin
J Antibiot , 73(9):650-654 (2020). - Beedessee, G., Kubota, T., Arimoto, A., Nishitsuji, K., Waller, R.F., Hisata, K., Yamasaki, S., Satoh, N., Kobayashi, J., Shoguchi, E.
Integrated omics unveil the secondary metabolic landscape of a basal dinoflagellate
BMC Biology, 18, Article number: 139 (2020) - Nogawa, T., Terai, A., Amagai, K., Hashimoto, J., Futamura, Y., Okano, A., Fujie, M., Satoh, N., Ikeda, H., Shin-Ya, K., Osada, H., Takahashi, S.
Heterologous Expression of the Biosynthetic Gene Cluster for Verticilactam and Identification of Analogues.
J Nat Prod. doi: 10.1021/acs.jnatprod.0c00755.
5. Seminar
No seminars in FY2020 due to COVID-2019.



