FY2016 Annual Report

Abstract
The genome contains all the genetic information of a given organism. Decoding the genome therefore provides the molecular basis for understanding every biological phenomenon. Since the turn of the 21st century, genomes of various metazoans have been sequenced, and consequently studies progressed efficiently in the fields of developmental biology, evolutionary biology, and environmental biology. The objective of the Marine Genomic Unit (MGU) is to pursue research in the realm of genome-based biological sciences by decoding genomes of target marine organisms (mainly invertebrates) so as to comprehensively elucidate the molecular mechanisms underlying (a) development and evolution of marine invertebrates, (b) environmental biology of corals, and (c) specific functions of marine organisms. We have decoded genome of the coral Acroporal digitifera in 2011 and the zooxanthella (coral-symbiotic brown algae) Symbiodinium minutum in 2013, respectively. OIST-MGU is therefore a lab in which the genomes of both coral and Symbiodinium have been sequenced. In addition, we have decoded genomes of the pearl oyster Pinctada fucata in 2012 and an anhydrobiotic midge with an extreme desiccation-tolerance ability in 2014. We are proud of deciphering two metazoan genomes in 2015; one is hemichordate genomes and the other is a brachiopod genome. In this year, 2016, we decoded a draft genome of the brown alga, Cladosiphon okamuranus, for a platform for future studies of “mozuku” biology. Also several advance was evident in other fields of MGU research. Main results shall be reported below.
1. Staff
- Professor Noriyuki Satoh
- Group Leaders
- Eiichi Shoguchi
- Chuya Shinzato (~1/31, '17)
- Staff Scientists
- Keisuke Nakashima
- Konstantin Khalturin
- Postdoctoral Scholars
- Ken Maeda
- Takeshi Takeuchi (7/1, '16 ~)
- Fuki Gyoja
- Yuuri Yasuoka
- Koki Nishitsuji
- Yuna Zayasu
- Asuka Arimoto
- Students
- Kenneth Baughman
- Keita Ikegami
- Yi-Jyun Luo
- Tsai-Ming Lu
- Girish BeeDessee
- Yafei Mao
- Technical Staffs
- Kanako Hisata
- Sakura Kikuchi
- Seiya Kitanobo (~11/1, '16)
- Research Assistants
- Yuki Yasuoka
- Yu-Pei Huang
- Yoshie Nishitsuji
- Research Administrators
- Ms. Shoko Yamakawa
- Ms. Tomomi Teruya
3. Activities and Findings
3.1 Developmental and evolutionary genomics
3.1.1 Chordate Origins and Evolution
We are interested in the origins and evolution of chordates. Chordates consist of three taxa, cephalochordates, urochordates, and vertebrates. Chordates are also a group of deuterostomes together with echinoderms and hemichordates. We believe that genome information provides basis for future studies of the origins and evolution of chordates. With such aim, we decoded genomes of a urochordate, Ciona intestinalis, in 2002 (Science 298: 2157-2167), a cephalochordate, Branchiostoma floridae, in 2008 (Nature 453: 1064-1071), two acorn worms, Saccoglossus kowalevskii and Ptychodera flava, in 2015 (Nature 527: 459-465), and a crown-of-thorns starfish, Acanthaster planci, in 2016 (unpublished). Since many vertebrate genomes have been reported, we have now obtained genomic information of all of the five deuterostome taxa.
N. Satoh has published a book entitled “Chordate Origins and Evolution” (Academic Press, 2016), in which he discussed how we can interpret developmental, molecular, and genome scientific mechanisms involved in the origins and evolution of chordates (Fig. 1). His major idea is that change or modification of larval form, namely larvae with cilia-swing-based swimming in ambulacrarians to larvae with tail-beat-based swimming in chordates is the key developmental event involved in the origin and evolution of chordates. This idea stands contrast to the “dorso-ventral axis inversion” hypothesis, and provides cues for further discussion of this, one of the most intriguing subjects of evolutionary developmental biology.

Figure 1: A book “Chordate Origins and Evolution” by N. Satoh (2016).
3.1.2 Brachyury in coral body plan
Brachyury is a member of the T-box family transcription factor genes. We have proposed the two-domain hypothesis of the gene expression and function; namely primary domain is at blastopore and stomodeum, which looks common to metazoans, and secondary domain is at notochord, which looks specific to chordates. In this context, its role in vertebrate mesoderm formation has been intensively studied. However, its fundamental function near the blastopore is poorly understood in other phyla. Cnidarians(diploblasts) are basal metazoans that are important for understanding evolution of metazoan body plans, since they lack mesoderm. We (Yasuoka et al., Current Biology 26: 1-8, 2016) carried out the first gene function analysis in corals. We showed that Brachyury maintains the ectoderm-endoderm border in coral embryos, which is directly activated by Wnt/b-catenin signaling in the ectoderm of coral embryos, indicating that the regulatory axis from Wnt/b-catenin signaling to Brachyury is highly conserved among eumetazoans (Fig. 2). Loss-of-function analysis demonstrated that Brachyury is required for pharynx formation but not for gastrulation movement. Genome wide transcriptome analysis demonstrated that genes positively regulated by Brachyury are expressed in the ectoderm of Acropora gastrulae, while negatively regulated genes are in endoderm. Therefore, germ layer demarcation around the blastopore appears to be the evolutionarily conserved role of Brachyury during gastrulation. Compared with Brachyury functions in vertebrate mesoderm-ectoderm and mesoderm-endoderm demarcation, the results suggest that the vertebrate-type mesoderm may have originated from Brachyury-expressing ectoderm adjacent to endoderm.
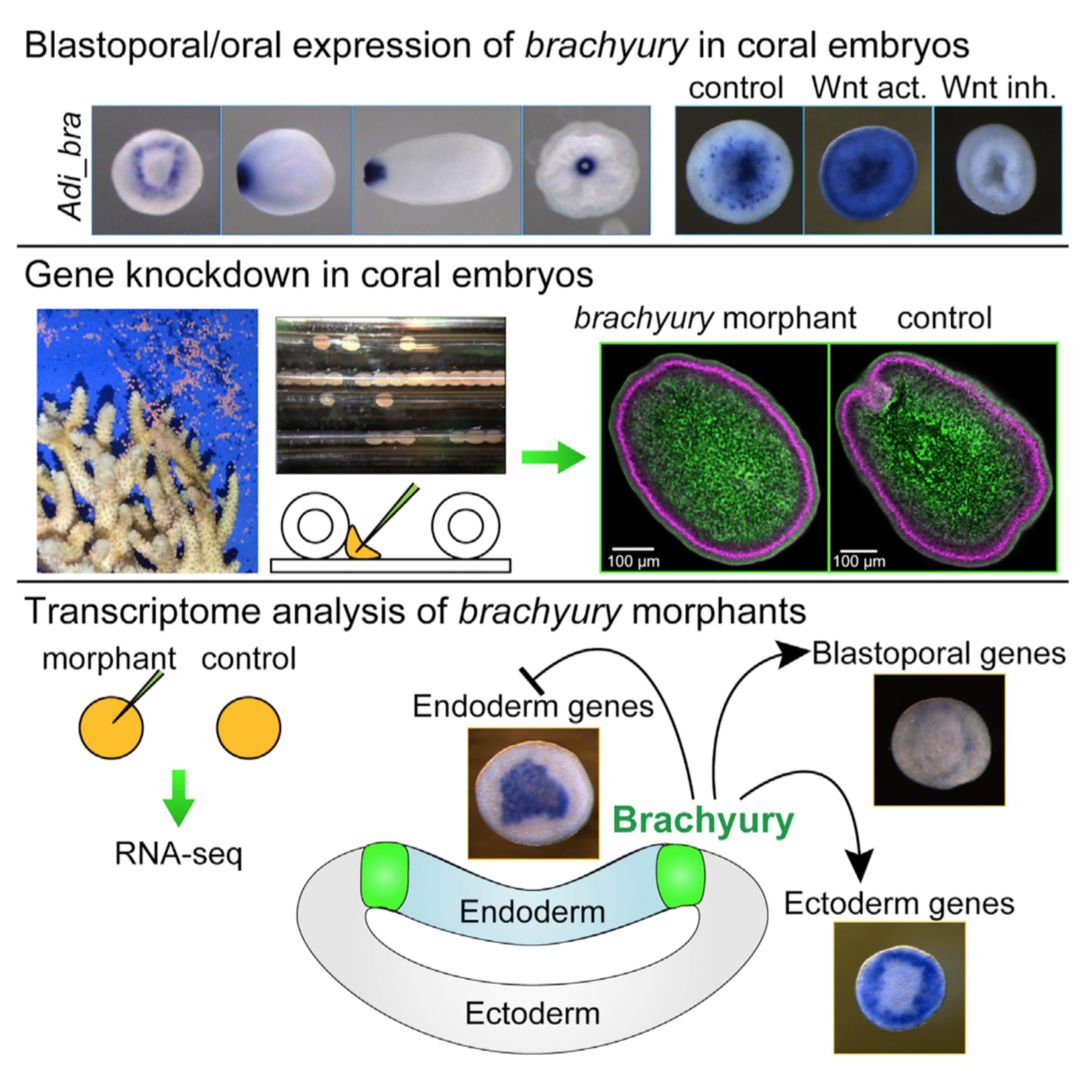
Figure 2
3.2 Environmental genomics
The coral reefs of the Okinawa islands are amongst the most biologically diverse ecosystems in the world. The key organisms in their establishment, the scleractinian corals, increasingly face a range of human-caused challenges including seawater temperature rises, ocean acidification and predation by COTS. To understand better the molecular mechanisms underlying coral biology, we succeeded in deciphering the 420-Mbp-long genome of the coral Acropora digitifera in 2011. Following a global coral bleaching event in 1998, Acropora corals surrounding most of Okinawa island were devastated, although they are now gradually recovering. In addition, a coral conservation/preservation project has been led by Okinawa Prefecture. Artificial transplantation of corals obtained by both sexual and asexual reproduction is one of the methods for the preservation, in which genetic diversity of corals should be kept in mind. MGU has establish a method to identify Acropora coral individuals as follows (Shinzato et al., Front. Marine Sci. 1: 11, 2014). The genus Acropora (Scleractinia, Acroporidae) is one of the most-wide spread coral genera, comprising the largest number of extant species among scleractinian (reef-building) corals. Molecular phylogenetic studies have suggested that A. tenuis belongs to the most basal clade (clade I) while A. digitifera belongs to a derived clade (clade IV). In order to develop microsatellite markers that would be useful for most Acropora species, we sequenced the genomic DNA of A. tenuis, using a next generation sequencer, and designed primer sets that amplify microsatellite loci. Afterward we selected primer pairs with perfectly matched nucleotide sequences from which at least one primer was uniquely mapped to the A. digitifera genome. Fourteen microsatellite markers showed non-significant departure from Hardy-Weinberg equilibrium in both A. tenuis and A. digitifera. Thus these markers could be used for wide range of species and may provide powerful tools for population genetics studies and conservation of Acropora corals.
In this year, 2016, using the method mentioned above, we challenged the population structure and genetic diversity of Acropora tenuis, one of the dominant stony coral species in the Pacific, using 13 microsatellite markers to investigate 298 colonies from 15 locations across the Nansei Islands in southwestern Japan (Zayasu et al., Ecol. Evol. 2016) (Fig. 3). Genetic diversity was not significant among sampling locations, even in possibly peripheral locations. In addition, our results showed that there are at least two populations of A. tenuis in the study area. The level of genetic differentiation between these populations was relatively low, but significant between many pairs of sampling locations. Directions of gene flow, which were estimated using a coalescence-based approach, suggest that gene flow not only occurs from south to north, but also from north to south in various locations. Consequently, the Yaeyama Islands and the Amami Islands are potential northern and southern sources of corals. On the other hand, the Miyako Islands and west central Okinawa Island are potential sink populations. The Kerama Islands and the vicinity of Taketomi Island are potential contact points of genetic subdivision of coral populations in the Nansei Islands. Namely, genetic population structure of A. tenuis in the Nansei Islands is more complex than previously thought. These cryptic populations are very important for preserving genetic diversity and should be maintained.
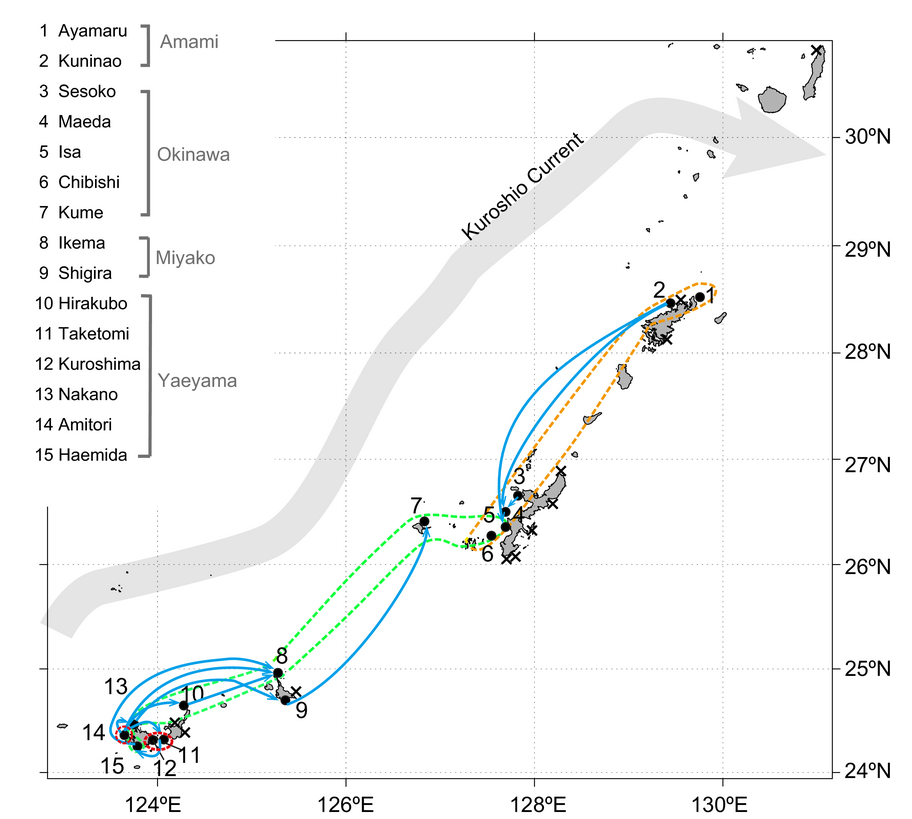
3.3 Functional genomics
In MGU functional genomics field, we wish to decode and analyze genomes of marine organisms, which are used for fishery and/or pharmaceutical production. The brown alga, Cladosiphon okamuranus (Okinawa mozuku), is economically one of the most important edible seaweeds, and is cultivated for market primarily in Okinawa, Japan. C. okamuranus constitutes a significant source of fucoidan, which has various physiological and biological activities. To facilitate studies of seaweed biology, we decoded the draft genome of C. okamuranus S-strain in collaboration Okinawa Prefectural Fisheries Research and Extension Center (Nishitsuji et al., DNA Research 23: 561-570, 2016). The genome size of C. okamuranus was estimated as 140 Mbp, smaller than genomes of two other brown algae, Ectocarpus siliculosus and Saccharina japonica. Together with transcriptomic data, we estimated that the C. okamuranus genome contains 13,640 protein-coding genes, approximately 94% of which have been confirmed with corresponding mRNAs. Comparisons with the E. siliculosus genome identified a set of C. okamuranus genes that encode enzymes involved in biosynthetic pathways for sulfated fucans (Fig. 4) and alginate biosynthesis. In addition, we identified C. okamuranus genes for enzymes involved in phlorotannin biosynthesis. The decoding of the Cladosiphon okamuranus genome provides a platform for future studies of mozuku biology. Further genome-wide examination of genes for enzymes involved in phlorotannin biosynthesis is now ongoing.
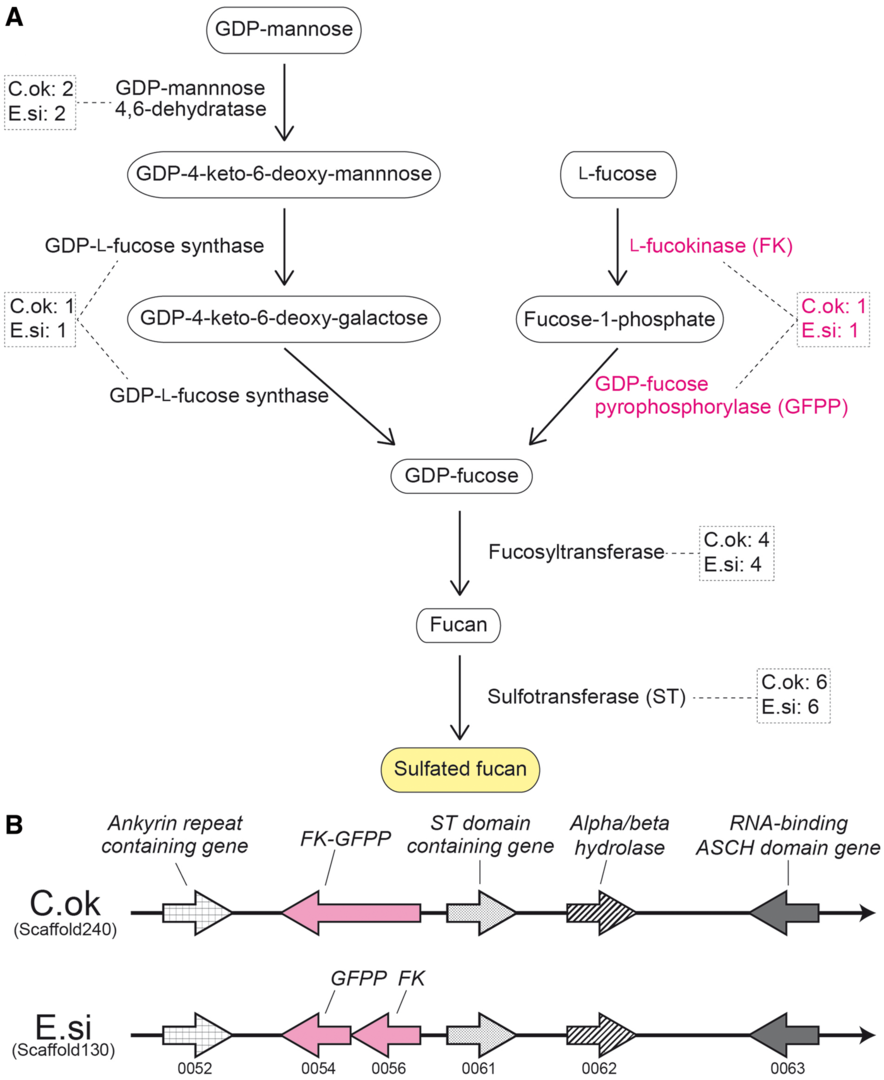
4. Publications
4.1 Journals
(a) Developmental and Evolutionary Genomics
- Satoh, N.
Chordate Origins and Evolution
Elsevier/Academic Press, 206 pages (2016)
- Yasuoka, Y., Shinzato, C., Satoh, N.
The mesoderm-forming gene brachyury regulates ectoderm-endoderm demarcation in the coral Acropora digitifera
Current Biology pii: S0960-9822(16)30923-X. doi: 10.1016/j.cub.2016.08.011. (2016)

- Satoh, N.
Two decades of ascidian developmental biology: A personal research story.
Curr. Topics Dev. Biol. 117:289-300 (2016)

- Satoh, N.
"Evolutionary Origin of Notochord" in "Chordomas.
Ed, Pamir, M.N. Thieme Publishers. New York. (2016) - Takeuchi, T., Yamada, L., Shinzato, C., Sawada, H., Satoh, N.
Stepwise evolution of coral biomineralization revealed with genome-wide proteomics and transcriptomics
PLOS ONE 11(6): e0156424. DOI:10.1371/journal.pone.0156424 (2016)

- Marin F, Bundeleva I, Takeuchi T, Immel F, Medakovic D
Organic matrices in metazoan calcium carbonate skeletons: Composition, functions, evolution
Journal of Structural Biology doi:10.1016/j.jsb.2016.04.006 (2016)

- Kodama H, Miyata Y, Kuwajima M, Izuchi R, Kobayashi A, Gyoja F, Onuma TA, Kumano G, Nishida H.
Redundant mechanisms are involved in suppression of default cell fates during embryonic mesenchyme and notochord induction in ascidians.
Developmental Biology 416: 162-172. doi: 10.1016/j.ydbio.2016.05.033.(2016)

- Suzuki, M., Mori, T., Satoh, N.
The Ciona intestinalis cleavage clock is independent of DNA methylation.
Genomics pii: S0888-7543(16)30091-X. doi: 10.1016/j.ygeno.2016.10.001. (2016)
- Session, A.M., Uno, Y., Kwon, T., Chapman, J.A., Toyoda, A., Takahashi, S., Fukui, A., Hikosaka, A., Suzuki, A., Kondo, M., van Heeringen, S.J., Quigley, I., Heinz, S., Ogino, H., Ochi, H., Hellsten, U., Lyons, J.B., Simakov, O., Putnam, N., Stites, J., Kuroki, Y., Tanaka, T., Michiue, T., Watanabe, M., Bogdanovic, O., Lister, R., Georgiou, G., Paranjpe, S.S., van Kruijsbergen, I., Shu, S., Carlson, J., Kinoshita, T., Ohta, Y., Mawaribuchi, S., Jenkins, J., Grimwood, J., Schmutz, J., Mitros, T., Mozaffari, S.V., Suzuki, Y., Haramoto, Y., Yamamoto, T.S., Takagi, C., Heald, R., Miller, K., Haudenschild, C., Kitzman, J., Nakayama, T., Izutsu, Y., Robert, J., Fortriede, J., Burns, K., Lotay, V., Karimi, K., Yasuoka, Y., Dichmann, D.S., Flajnik, M.F., Houston, D.W., Shendure, J., DuPasquier, L., Vize, P.D., Zorn, A.M., Ito, M., Marcotte, E.M., Wallingford, J.B., Ito, Y., Asashima,M., Ueno, N., Matsuda, Y., Veenstra, G.J.C., Fujiyama, A., Harland, R.M., Taira, M., Rokhsar, D.S.
Genome evolution in the allotetraploid frog Xenopus laevis
Nature 538, 336–343.

- Watanabe, M., Yasuoka, Y., Mawaribuchi, S., Kuretani, A., Ito, M., Kondo, M., Ochi, H., Ogino, H., Fukui, A., Taira, M., Kinoshita, T.
Conservatism and variability of gene expression profiles among homeologous transcription factors in Xenopus laevis.
Dev Biol. pii: S0012-1606(16)30053-7. doi: 10.1016/j.ydbio.2016.09.017.
(b) Environmental Genomics
- Zayasu, Y., Shinzato, C.
Hope for coral reef rehabilitation: massive synchronous spawning by outplanted corals in Okinawa, Japan.
Coral Reefs, 35(4), 1295. DOI 10.1007/s00338-016-1463-7 (2016)
- Zayasu, Y., Nakajima, Y., Sakai, K., Suzuki, G., Satoh, N., Shinzato, C.
Unexpectedly complex gradation of coral population structure in the Nansei Islands, Japan.
Ecology and Evolution. on line. 10.1002/ece3.2296 (2016).

- Hosokawa, T., Ishii, Y., Nikoh, N., Fujie, M., Satoh, N., Fukatsu, T.
Obligate bacterial mutualists evolving from environmental bacteria in natural insect populations.
Nature Microbiology, 1:15011 (2016)

- Detree, C., Chabenat, A., Lallier, F.H., Satoh, N., Shoguchi, E., Tanguy, A., Mary, J.
Multiple I-Type Lysozymes in the Hydrothermal Vent Mussel Bathymodiolus azoricus and Their Role in Symbiotic Plasticity.
PLoS One 11(2):e0148988. doi: 10.1371/journal.pone.0148988. (2016)
- Nakajima, Y., Zayasu, Y., Shinzato, C., Satoh, N., Mitarai. S.
Genetic differentiation and connectivity of morphological types of the broadcast-spawning coral Galaxea fascicularis in the Nansei Islands, Japan
Ecology and Evolution, 6:1457-1469 (2016)

- Mohamed AR, Cumbo V, Harii S, Shinzato C, Chan CX, Ragan MA, Bourne DG, Willis BL, Ball EE, Satoh N, Miller DJ.
The transcriptomic response of the coral Acropora digitifera to a competent Symbiodinium strain: the symbiosome as an arrested early phagosome.
Mol Ecol. doi: 10.1111/mec.13659. (2016)

- Nakajima Y, Shinzato C, Khalturina M, Nakamura M, Watanabe H, Satoh N, Mitarai S.
The mitochondrial genome sequence of a deep-sea, hydrothermal vent limpet, Lepetodrilus nux, presents a novel vetigastropod gene arrangement.
Mar Genomics. pii: S1874-7787(16)30030-7. doi: 10.1016/j.margen.2016.04.005. (2016)

- Bhattacharya, D., Agrawal, S., Aranda, M., Baumgarten, S., Belcaid, M., Drake, J.L., Erwin, D., Foret, S., Gates, R.D., Gruber, D.F., Kamel, B., Lesser, M.P., Levy, O., Liew, Y.J., MacManes, M., Mass, T., Medina, M., Mehr, S., Meyer, E., Price, D.C., Putnam, H.M., Qiu, H., Shinzato, C., Shoguchi, E., Stokes, A.J., Tambutté, S., Tchernov, D., Voolstra, C.R., Wagner, N., Walker, C.W., Weber, A.P.M., Weis, V., Zelzion, E., Zoccola, D., Falkowski, P.G.
Comparative genomics explains the evolutionary success of reef-forming corals.
eLife 2016;5:e13288, DOI: http://dx.doi.org/10.7554/eLife.13288 (2016)

- Arrigoni, R., Benzoni, F., Huang, D., Fukami, H., Chen, C.A., Berumen, M.L., Hoogenboom, M., Thomson, D.P., Hoeksema, B.W., Budd, A.F., Zayasu, Y., Terraneo, T. I., Kitano, Y.F., Baird, A.H.
When forms meet genes: revision of the scleractinian genera Micromussa and Homophyllia (Lobophylliidae) with a description of two new species and one new genus.
Contributions to Zoology, 85 (4): 387-422. (2016)
- Omori, M., Higa, Y., Shinzato, C., Zayasu, Y., Nagata, T., Nakamura, R., Yokokura, A., Janadou, S.
Development of active restoration methodologies for coral reefs using asexual reproduction in Okinawa, Japan.
Proceeding of the 13th International Coral Reef Symposium: 369–387 (2016)
(c) Functional Genomics
- Nishitsuji, K., Arimoto, A., Iwai, K., Sudo, Y., Hisata, K., Fujie, M., Arakaki, N., Kushiro, T., Konishi, T., Shinzato, C., Satoh, N., Shoguchi, E.
A draft genome of the brown alga, Cladosiphon okamuranus, S-strain: a platform for future studies of 'mozuku' biology.
DNA research pii: dsw039 (2016)

- Takeuchi, T., Koyanagi, R., Gyoja, F., Kanda, M., Hisata, K., Fujie, M., Goto, H., Yamasaki, S., Nagai, K., Morino, Y., Miyamoto, H., Endo, K., Endo, H., Nagasawa, H., Kinoshita., S., Asakawa, S., Watabe, S., Satoh, N., Kawashima, T.
Bivalve-specific gene expansion in the pearl oyster genome: Implications of adaptation to a sessile lifestyle
Zoological Letters, 2:3 (2016)

(d) Other Fields
- Katsuyama Y, Sone K, Satou R, Izumikawa M, Takagi M, Fujie M, Satoh N, Shin-Ya K, Ohnishi Y.
Involvement of the Baeyer-Villiger Monooxygenase IfnQ in the Biosynthesis of Isofuranonaphthoquinone Scaffold of JBIR-76 and -77.
Chembiochem. 2;17(11):1021-8. doi: 10.1002/cbic.201600095. (2016)
- Du D, Katsuyama Y, Onaka H, Fujie M, Satoh N, Shin-Ya K, Ohnishi Y.
Production of a Novel Amide-Containing Polyene by Activating a Cryptic Biosynthetic Gene Cluster in Streptomyces sp. MSC090213JE08.
Chembiochem. 3;17(15):1464-71. doi: 10.1002/cbic.201600167. (2016)
- Iida, M., Kondo, M., Tabouret, H., Maeda, K., Pécheyran, C., Hagiwara, A., Keith, P., Tachihara, K.
Specific gravity and migratory patterns of amphidromous gobioid fish from Okinawa Island, Japan Journal of Experimental Marine Biology and Ecology, 486: 160–169. (2016)



