FY2015 Annual Report

Abstract
The genome contains all the genetic information of a given organism. Decoding the genome therefore provides the molecular basis for understanding every biological phenomenon. Since the turn of the 21st century, genomes of various metazoans have been sequenced, and consequently studies progressed efficiently in the fields of evolutionary biology, developmental biology, and environmental biology. The objective of the Marine Genomic Unit (MGU) is to pursue research in the realm of genome-based biological sciences by decoding the genomes of target marine invertebrates so as to comprehensively elucidate the molecular mechanisms underlying (a) development and evolution of marine invertebrates, (b) environmental responses of corals, and (c) specific functions of marine organisms. In 2011 we have decoded genome of the coral Acroporal digitifera, and in 2013 we succeeded in decoding the genome of a zooxanthella (coral-symbiotic brown algae), Symbiodinium minutum. OIST-MGU is therefore a lab in which the genomes of both coral and Symbiodinium have been sequenced. In addition, we have decoded genomes of the pearl oyster Pinctada fucata in 2012 and an anhydrobiotic midge with an extreme desiccation-tolerance ability in 2014. We are proud of deciphering two metazoan genomes in fiscal year 2015; one is hemichordate genomes and the other is a brachiopod genome. In this year, we also advanced population genetics of Acropora corals in the Okinawa islands.
(a) Developmental and evolutionary genomics:
We are interested in the origins and evolution of chordates. Chordates consist of three taxa, cephalochordates, urochordates, and vertebrates. Chordates are also a group of deuterostomes together with echinoderms and hemichordates. We believe that genome information provides basis for future studies of the origins and evolution of chordates. With such aim, we decoded genomes of a urochordate, Ciona intestinalis, in 2002 (Science 298: 2157-2167) and a cephalochordate, Branchiostoma floridae, in 2008 (Nature 453: 1064-1071). Since the sea urchin genome sequence consortium decoded the Strongylocentrotus purpuratus genome in 2006, hemichordates are deuterostomes of which genome remains to be sequenced. In collaboration with researchers of UC Berkeley and other University, we have finally decoded the genome of two acorn worms, Saccoglossus kowalevskii and Ptychodera flava. Thus, we have now obtained genomic information of all of the five deuterostome taxa. The hemichordate genome tells us what deuterostomes are. We also deciphered a brachiopod genome.
(b) Environmental genomics:
The coral reefs of the Okinawa islands are amongst the most biologically diverse ecosystems in the world. The key organisms in their establishment, the scleractinian corals, increasingly face a range of human-caused challenges including seawater temperature rises and ocean acidification. Following a global coral bleaching event in 1998, Acropora corals surrounding most of Okinawa island (OI) were devastated, although they are now gradually recovering. In contrast, the Kerama Islands (KIs) only 30 km west of OI, have continuously hosted a great variety of healthy corals. We deciphered the 420-Mbp-long genome of the coral Acropora digitifera in 2011. This year, using the genomic information, we analyzed at single nucleotid polymorphism (SNP) level how the OI population has recovered, and we obtained interesting results.
(c) Functional genomics:
Dinoflagellates are unicellular marine and freshwater eukaryotes. They possess large nuclear genomes (1.5–245 gigabases) and produce structurally unique and biologically active polyketide secondary metabolites. Aforementioned, we have decoded the genome of a coral symbiont, Symbiodinium minutum nuclear genome, for the first time in the word (Curr. Biol. 23: 1399-1408, 2013). Together with transcriptomic data, this year we have investigated the assembled S. minutum genome and identified 25 candidate polyketide synthase (PKS) genes that encode proteins with mono- and multifunctional domains. Predicted proteins retain functionally important amino acids in the catalytic ketosynthase (KS) domain. Molecular phylogenetic analyses of KS domains form a clade in which S. minutum domains cluster within the protist Type I PKS clade.
1. Staff
- Group Leaders
- Eiichi Shoguchi
- Chuya Shinzato
- Staff Scientists
- Mayuko Hamada
- Keisuke Nakashima
- Konstantin Khalturin
- Postdoctoral Scholars
- Ken Maeda
- Takeshi Takeuchi
- Fuki Gyoja
- Yuuri Yasuoka (JSPS, PD Fellow)
- Koki Nishitsuji
- Yuna Zayasu
- Asuka Arimoto
- Students
- Kenneth Baughman
- Keita Ikegami
- Yi-Jyun Luo
- Tsai-Ming Lu
- Girish BeeDessee
- Yafei Mao
- Technical Staffs
- Kanako Hisata
- Sakura Kikuchi
- Mariia Khalturina
- Yoshie Nishitsuji
- Yu-Pei Huang
- Research Assistants
- Ms. Yuki Yasuoka
- Research Administrators
- Ms. Shoko Yamakawa
- Ms. Tomomi Teruya
3. Activities and Findings
3.1 Developmental and evolutionary genomics
3.1.1 Hemichordate genomes
Acorn worms, also known as enteropneust (literally, ‘gut-breathing’) hemichordates, are marine invertebrates that share features with echinoderms and chordates. Together, these three phyla comprise the deuterostomes. In this year we report the draft genome sequences of two acorn worms, Saccoglossus kowalevskii and Ptychodera flava, both approximately 1-gigabase genome containing at least 18,556 and 19,270 genes, respectively (Nature 527: 459-465, 2015). By comparing them with diverse bilaterian genomes, we identify shared traits that were probably inherited from the last common deuterostome ancestor, and then explore evolutionary trajectories leading from this ancestor to hemichordates, echinoderms and chordates. The hemichordate genomes exhibit extensive conserved synteny with amphioxus and other bilaterians, and deeply conserved non-coding sequences that are candidates for conserved gene-regulatory elements. Notably, as shown in Fig. 1, hemichordates possess a deuterostome-specific genomic cluster of four ordered transcription factor genes (Nkx2.1, Nkx2.2, Pax1/9, and FoxA), the expression of which is associated with the development of pharyngeal ‘gill’ slits, the foremost morphological innovation of early deuterostomes, and is probably central to their filter-feeding lifestyle. Comparative analysis reveals numerous deuterostome-specific gene novelties, including genes found in deuterostomes and marine microbes, but not other animals. The putative functions of these genes can be linked to physiological, metabolic and developmental specializations of the filter-feeding ancestor.
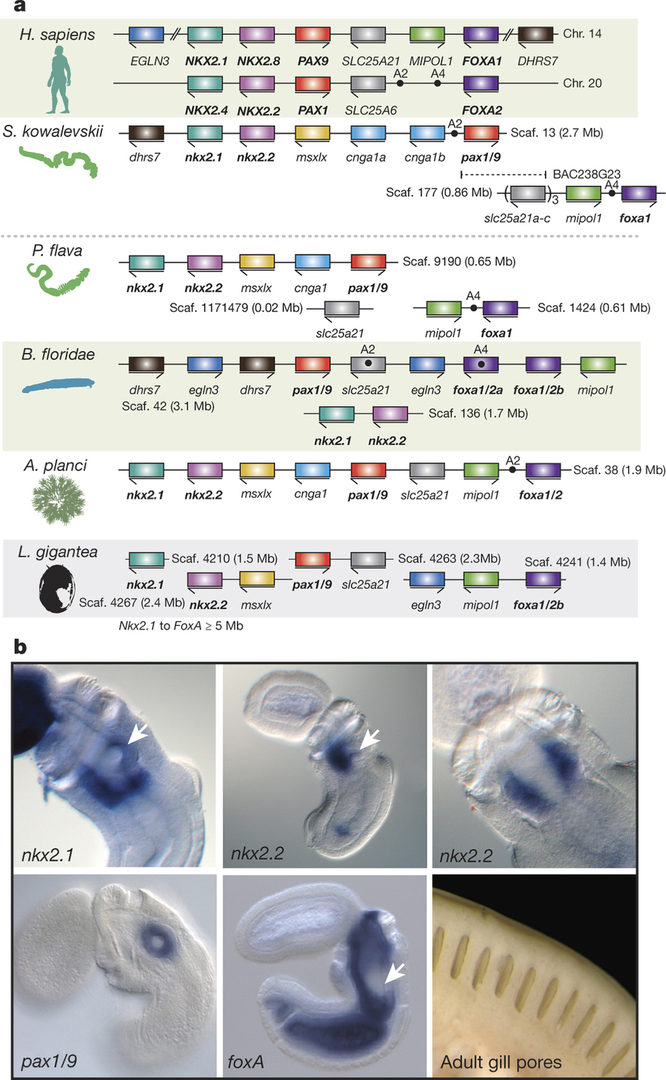
3.1.2 Brachiopod genome
Brachiopods are marine invertebrates with calcium phosphate or carbonate shells (Fig. 2).
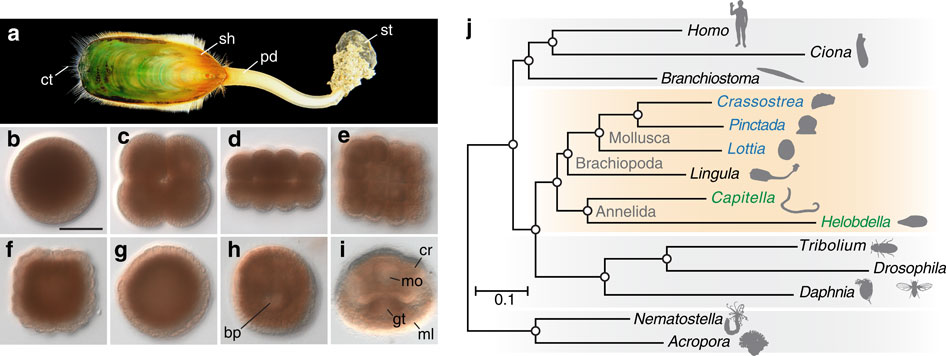
Figure 2. Deuterostomic development of the brachiopod, L. anatina, and its close relationship to molluscs. (a) Adult (shell length ~4–5 cm). (b–i) Embryogenesis: egg (b), embryos at 4-cell (c), 16-cell (d), 32-cell (e) and 128-cell stages (f), blastula (g), late gastrula (h) and 2-pair cirri larva (i). Scale bar, 50 μm. bp, blastopore; cr, cirri; ct, chaeta; gt, gut; ml, mantle lobe; mo, mouth; pd, pedicle; sh, shell; st, stone. (j) Phylogenetic position of Lingula among lophotrochozoans (orange box; molluscs are blue; annelids are green). The tree was constructed using the maximum likelihood method with 150 one-to-one orthologues (46,845 amino-acid positions) with LG+Γ4 model. Circles at all nodes indicate 100% bootstrap support.
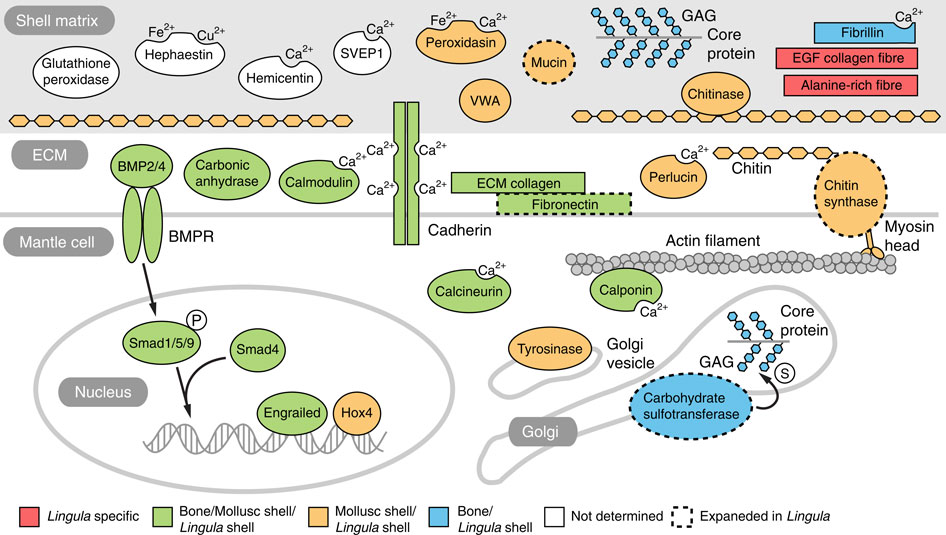
Abundant in the fossil record, Darwin first referred to lingulid brachiopods as ‘living fossils,’ because their shell morphology has changed little since the Silurian. The evolutionary origins of lingulid brachiopods and their calcium phosphate shells have been obscure. This year, we decode the 425-Mb genome of Lingula anatina to gain insights into brachiopod evolution (Nature Communication 6: 8301, 2015). Comprehensive phylogenomic analyses place Lingula close to molluscs, but distant from annelids. The Lingula gene number has increased to ~34,000 by extensive expansion of gene families. Although Lingula and vertebrates have superficially similar hard tissue components, our genomic, transcriptomic and proteomic analyses show that Lingula lacks genes involved in bone formation, indicating an independent origin of their phosphate biominerals (Fig. 3). Several genes involved in Lingula shell formation are shared by molluscs. However, Lingula has independently undergone domain combinations to produce shell matrix collagens with EGF domains and carries lineage-specific shell matrix proteins. Gene family expansion, domain shuffling and co-option of genes appear to be the genomic background of Lingula’s unique biomineralization. This Lingula genome provides resources for further studies of lophotrochozoan evolution.
3.2 Environmental genomics
The coral reefs of the Okinawa islands are amongst the most biologically diverse ecosystems in the world. The key organisms in their establishment, the scleractinian corals, increasingly face a range of human-caused challenges including seawater temperature rises and ocean acidification. To understand better the molecular mechanisms underlying coral biology, we succeeded in deciphering the 420-Mbp-long genome of the coral Acropora digitifera in 2011. Following a global coral bleaching event in 1998, Acropora corals surrounding most of Okinawa island (OI) were devastated, although they are now gradually recovering. In contrast, the Kerama Islands (KIs) only 30 km west of OI, have continuously hosted a great variety of healthy corals. Taking advantage of the decoded Acropora digitifera genome and using genome-wide SNP analyses, we clarified Acropora population structure in the southern Ryukyu Archipelago (sRA) (Sci. Reports 5: 18211, 2015).
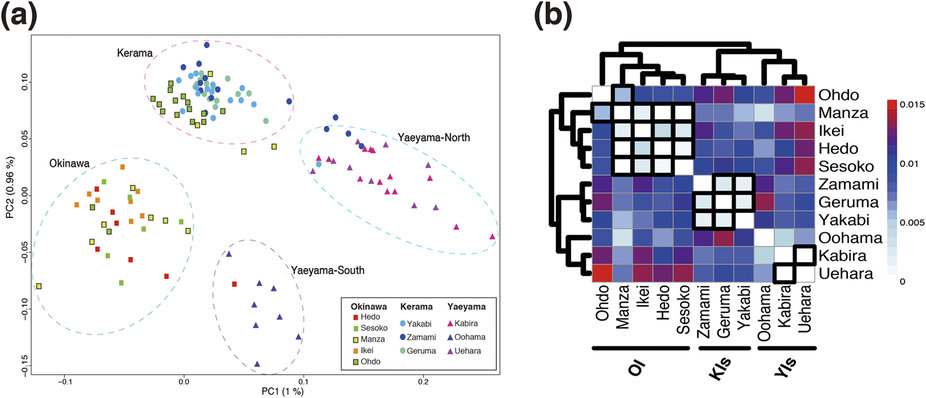
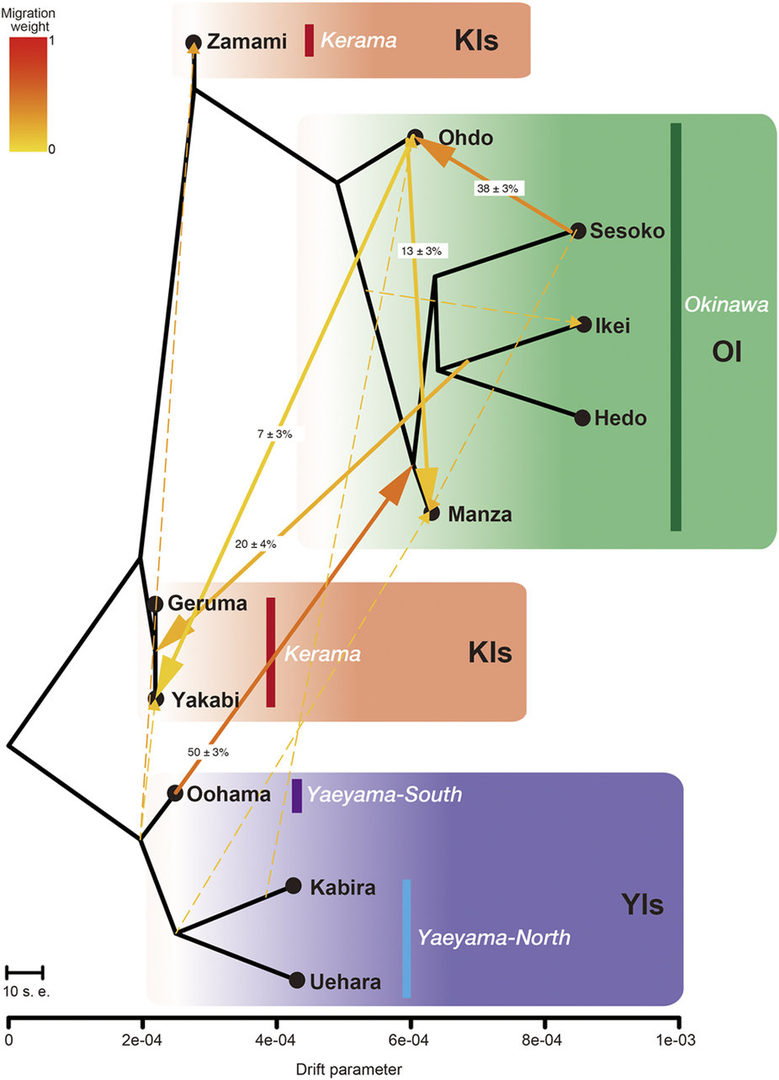
As shown in Fig. 4, despite small genetic distances, we identified distinct clusters corresponding to specific island groups, suggesting infrequent long-distance dispersal within the sRA. Although the KIs were believed to supply coral larvae to OI, admixture analyses showed that such dispersal is much more limited than previously realized, indicating independent recovery of OI coral populations and the necessity of local conservation efforts for each region. We detected strong historical migration from the Yaeyama Islands (YIs) to OI, and suggest that the YIs are the original source of OI corals (Fig. 5). In addition, migration edges to the KIs suggest that they are a historical sink population in the sRA, resulting in high diversity. This population genomics study provides the highest resolution data to date regarding coral population structure and history.
3.3 Functional genomics
Dinoflagellates are unicellular marine and freshwater eukaryotes. They possess large nuclear genomes (1.5–245 gigabases) and produce structurally unique and biologically active polyketide secondary metabolites. Aforementioned, we have decoded the genome of a coral symbionts, Symbiodinium minutum nuclear genome, for the first time in the word (Curr. Biol. 23: 1399-1408, 2013). Together with transcriptomic analyses, we have investigated the assembled S. minutum genome and identified 25 candidate polyketide synthase (PKS) genes that encode proteins with mono- and multifunctional domains (BMC Genomics 16: 2385-2391, 2015). Predicted proteins retain functionally important amino acids in the catalytic ketosynthase (KS) domain. Molecular phylogenetic analyses of KS domains form a clade in which S. minutum domains cluster within the protist Type I PKS clade with those of other dinoflagellates and other eukaryotes. Single-domain PKS genes are likely expanded in dinoflagellate lineage. Two PKS genes of bacterial origin are found in the S. minutum genome. Interestingly, the largest enzyme is likely expressed as a hybrid non-ribosomal peptide synthetase-polyketide synthase (NRPS-PKS) assembly of 10,601 amino acids, containing NRPS and PKS modules and a thioesterase (TE) domain. We also found intron-rich genes with the minimal set of catalytic domains needed to produce polyketides. Ketosynthase (KS), acyltransferase (AT), and acyl carrier protein (ACP) along with other optional domains are present. Mapping of transcripts to the genome with the dinoflagellate-specific spliced leader sequence, supports expression of multifunctional PKS genes. Metabolite profiling of cultured S. minutum confirmed production of zooxanthellamide D, a polyhydroxy amide polyketide and other unknown polyketide secondary metabolites.
Aforementioned, this genomic survey demonstrates that S. minutum contains genes with the minimal set of catalytic domains needed to produce polyketides and provides evidence of the modular nature of Type I PKS, unlike monofunctional Type I PKS from other dinoflagellates. In addition, our study suggests that diversification of dinoflagellate PKS genes comprises dinoflagellate-specific PKS genes with single domains, multifunctional PKS genes with KS domains orthologous to those of other protists, and PKS genes of bacterial origin.
4. Publications
4.1 Journals
(a) Developmental and Evolutionary Genomics
- Simakov, O., Kawashima, T., Marlétaz, F., Jenkins, J., Koyanagi, R., Mitros, T., Hisata, K., Bredeson, J., Shoguchi, E., Gyoja, F., Yue, J.X., Chen, Y.C., Robert M. Freeman, R.M., Sasaki, A.,Hikosaka-Katayama, T., Sato, A., Fujie, M., Baughman, K.W., Levine, J., Paul Gonzalez, P., Cameron, C., Fritzenwanker, J.H., Pani, A.M., Goto, H., Kanda, M., Arakaki, N., Yamasaki, S., Qu, J., Cree, A., Ding, Y., Dinh, H.H., Dugan, S., Holder, M., Jhangiani, S.N., Kovar, C.L., Lee, S.L., Lewis, L.R., Morton, D., Nazareth, L.V., Okwuonu, G., Santibanez, J., Chen, R., Richards, S., Muzny, D.M., Gillis, A., Peshkin, L., Wu, M., Humphreys, T., Su, Y.H., Putnam, N.H., Schmutz, J., Fujiyama, A., Yu, J.K., Tagawa, K., Worley, K.C., Gibbs, R.A., Kirschner, M.W., Christopher J. Lowe, C.J., Satoh, N., Rokhsar, D.S., Gerhart, J.
Hemichordate genomes and deuterostome origins
Nature 527(7579):459-65. (2015)

- Luo, Y.J., Takeuchi, T., Koyanagi, R., Yamada, L., Kanda, M., Khalturina, M., Fujie, M., Yamasaki, S., Endo, K., Satoh, N.
The Lingula genome provides insights into brachiopod evolution and the origin of phosphate biomineralization.
Nature Communications, 18;6:8301. doi: 10.1038/ncomms9301. (2015)

- Kawai, N., Ogura, Y., Ikuta, T., Saiga, H., Hamada, M., Sakuma, T., Yamamoto, T., Satoh, N., Sasakura, Y.
Hox10-regulated endodermal cell migration is essential for development of the ascidian intestine.
Developmental Biology, S0012-1606(15)00169-4. doi: 10.1016/j.ydbio.2015.03.018. (2015).
- Hamada, M., Goricki, S., Byerly, M.S., Satoh, N., Jeffery, W.R.
Evolution of the chordate regeneration blastema: Differential gene expression and conserved role of notch signaling during siphon regeneration in the ascidian Ciona
Developmental Biology 15;405(2):304-15. (2015)
- Kraus, J.E.M., Fredman, D., Wang, W., E., Khalturin, K., Technau, U.
Adoption of conserved developmental genes in development and origin of the medusa body plan.
EvoDevo, 6:23 (2015)
- Luo, Y.J., Satoh, N., Endo, K.
Mitochondrial gene order variation in the brachiopod Lingula anatina and its implications for mitochondrial evolution in lophotrochozoans.
Marine Genomics, pii: S1874-7787(15)30021-0. doi: 10.1016/j.margen.2015.08.005. (2015)

- Alié, A., Hayashi, T., Sugimura, I., Manuel, M., Sugano, W., Mano, A., Satoh, N., Agata, K., Funayama, N.
The ancestral gene repertoire of animal stem cells
PNAS 112(51):E7093-100 doi: 10.1073/pnas.1514789112 (2015) online

- Satoh, N.
Two decades of ascidian developmental biology: A personal research story.
Curr. Topics Dev. Biol. 117:289-300 (2016)

(b) Environmental Genomics
- Maruyama, S., Shoguchi, E., Satoh, N., Minagawa, J.
Diversification of light harvesting complex gene family via intra- and intergenic duplications in the coral symbiotic alga Symbiodinium.
PLoS One 10(3):e0119406. (2015)
- Nakajima, Y., Shinzato, C., Satoh, N., Mitarai S.
Novel Polymorphic Microsatellite Markers Reveal Genetic Differentiation between Two Sympatric Types of Galaxea fascicularis.
PLoS One. 10(7):e0130176. (2015)
- Shoguchi, E., Shinzato, C., Hisata, K., Satoh, N., Mungpakdee, S.
The Large Mitochondrial Genome of Symbiodinium minutum Reveals Conserved Noncoding Sequences between Dinoflagellates and Apicomplexans..
Genome Biol Evol. 20;7(8):2237-44. (2015)
- Sato, A., Kawashima, T., Fujie, M., Hughes, S., Satoh, N., Shimeld, S.
Molecular basis of canalization in an ascidian species complex adapted to different thermal conditions.
Science Reports 18;5:16717 (2015)
- Shinzato, C., Mungpakdee, S., Arakaki, N., Satoh, N.
Genome-wide SNP analysis explains coral diversity and recovery in the Ryukyu Archipelago
Scientific Reports 5, Article number: 18211 (2015) doi:10.1038/srep18211

- Sayavedra, L., Kleiner, M., Ponnudurai, R., Wetzel, S., Pelletier, E., Barbe, V., Satoh, N., Shoguchi, E., Fink, D., Breusing, C., Reusch, T.B., Rosenstiel, P., Schilhabel, M.B., Becher, D., Schweder, T., Markert, S., Dubilier, N, Petersen, J.M.
Abundant toxin-related genes in the genomes of beneficial symbionts from deep-sea hydrothermal vent mussels.
Elife. 15;4. pii: e07966. doi: 10.7554/eLife.07966. (2015).
- Hosokawa, T., Ishii, Y., Nikoh, N., Fujie, M., Satoh, N., Fukatsu, T.
Obligate bacterial mutualists evolving from environmental bacteria in natural insect populations.
Nature Microbiology, 1:15011 (2016)
- Detree, C., Chabenat, A., Lallier, F.H., Satoh, N., Shoguchi, E., Tanguy, A., Mary, J.
Multiple I-Type Lysozymes in the Hydrothermal Vent Mussel Bathymodiolus azoricus and Their Role in Symbiotic Plasticity.
PLoS One 11(2):e0148988. doi: 10.1371/journal.pone.0148988. (2016)
- Nakajima, Y., Zayasu, Y., Shinzato, C., Satoh, N., Mitarai. S.
Genetic differentiation and connectivity of morphological types of the broadcast-spawning coral Galaxea fascicularis in the Nansei Islands, Japan
Ecology and Evolution, 6:1457-1469 (2016)

(c) Functional genomics
- Shin-ya, K., Izumikawa, M., Kozone, I., Hashimoto, J., Kagaya, N., Takagi, M., Koiwai, H., Komatsu, M., Fujie, M., Satoh, N., Ikeda, H.
Novel thioviridamide derivative-JBIR-140-heterologous expression of the gene cluster for thioviridamide biosynthesis.
Journal of Antibiotics, 68(8):533-6. doi: 10.1038/ja.2015.20. (2015).
- Ohno, S., Katsuyama, Y., Tajima, Y., Izumikawa, M., Takagi, M., Fujie, M., Satoh, N., Shin-ya, K., Yasuo Ohnishi, Y.
Identification and characterization of the streptazone E biosynthetic gene cluster in Streptomyces sp.
ChemBioChem 16(16):2385-91. (2015)
- Beedessee, G., Hisata, K., Roy, M.C., Satoh, N., Shoguchi, E.
Multifunctional polyketide synthase genes identified by genomic survey of the symbiotic dinoflagellate, Symbiodinium minutum.
BMC Genomics 16:941(2015)

- Ohno S, Katsuyama Y, Tajima Y, Izumikawa M, Takagi M, Fujie M, Satoh N, Shin-Ya K, Ohnishi Y.
Identification and Characterization of the Streptazone E Biosynthetic Gene Cluster in Streptomyces sp. MSC090213JE08.
Chembiochem. 16(16):2385-91. doi: 10.1002/cbic.201500317. (2015)

- Takeuchi, T., Koyanagi, R., Gyoja, F., Kanda, M., Hisata, K., Fujie, M., Goto, H., Yamasaki, S., Nagai, K., Morino, Y., Miyamoto, H., Endo, K., Endo, H., Nagasawa, H., Kinoshita., S., Asakawa, S., Watabe, S., Satoh, N., Kawashima, T.
Bivalve-specific gene expansion in the pearl oyster genome: Implications of adaptation to a sessile lifestyle
Zoological Letters, 2:3 (2016)

(d) Other Fields (Fish taxonomy and ecology)
- Maeda, K., Tran, H.D., Tan, H.H.
Discovery of a substantial continental population of the subfamily Sicydiinae (Gobioidei: Gobiidae) from Vietnam: Taxonomic revision of the genus Stiphodon from the western South China Sea.
Raffles Bulletin of Zoology 63: 246–258 (2015)
- Lord C., Maeda K., Keith P., Watanabe S.
Population structure of the Asian amphidromous Sicydiinae goby, Stiphodon percnopterygionus, inferred from mitochondrial COI sequences, with comments on larval dispersal in the northwest Pacific Ocean.
Life and Environment, 65: 63–71. (2015)
- Maeda, K., Palla, H.P.
A new species of the genus Stiphodon from Palawan, Philippines (Gobiidae: Sicydiinae)
Zootaxa 4018 (3): 381–395. (2015)

- Maeda, K., Iida, M., Kondo, M.
Diversity of larval dispersal strategies in freshwater and estuarine gobies
Mochioka, N., Kinoshita, I., Minami, T. (eds) Early life history of fishes, Kouseisha-Kouseikaku, Tokyo. pp. 89–101(2015) - Keith, P., Lord, C., and Maeda, K.
Indo-Pacific Sicydiine gobies: Biodiversity, life traits and conservation
Publisher: Société Française d’Ichtyologie, Paris (2015)
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- Luo, Y.-J., Takeuchi, T., Koyanagi, R., Yamada, L., Kanda, M., Khalturina, M., Fujie, M., Yamasaki, S., Endo, K., and Satoh, N. The brachiopod genome of Lingula anatina and the evolution of lophotrochozoans and biomineralization. The 7th International Brachiopod Congress. (Nanjing, China) (2015)
- Yasuoka, Y., Koyanagi, R., and Satoh, N. Subfunctionalization of two brachyury genes in Xenopus reveals Brachyury functions in the vertebrate notochord. The 48th Annual Meeting of the Japanese Society of Developmental Biologists. (Tsukuba International Congress Center, Tsukuba, Ibaraki) (2015)
- Khalturin, K., Shinzato, C., Khalturina, M., Fujie, M., Koyanagi, R., Goto, H., and Satoh, N. Life cycle regulation in Cnidaria and the origin of a jellyfish body plan. The 48th Annual Meeting of the Japanese Society of Developmental Biologists. (Tsukuba International Congress Center, Tsukuba, Ibaraki) (2015)
- Maeda, K., and Palla, H.P. An undescribed species of the genus Stiphodon from Palawan, Philippines. In GORI Meeting 2015 (Nagaragawa Ukai Museum). (2015)
- Iida, M., Kondo, M., Maeda, K., Tabouret, H., Keith, P., and Tachihara, K. Characteristics of amphidromous gobioid larvae in relation to larval retention and dispersal. 39th Annual Larval Fish Conference. (University of Vienna, Vienna, Austria) (2015)
- Luo, Y.-J. The brachiopod genome of Lingula anatina provides insight into the evolution of lophotrochozoans and calcium-phosphate-based biomineralization. Inaugural 2015 Meeting, Pan-American Society for Evolutionary Developmental Biology. (University of California Berkeley) (2015)
- Yasuoka, Y., Koyanagi, R., Yasuoka, Y., Shinzato, C., and Satoh, N. The vertebrate-mesodermal gene Brachyury regulates ectoderm-endoderm demarcation in a diploblastic animal, Acropora digitifera. In 17th Annual Meeting of Society of Evolutionary Studies, Japan (Chuo University Korakuen Campus, Tokyo ). (2015)
- Yasuoka, Y. Subfunctionalization of two brachyury genes in Xenopus In 1st meeting of Next Generation Amphibian Study (Okazaki Conference Center, National Institute of Natural Sciences, Aichi, Japan). (2015)
- Shoguchi, E. Comparative genomics of dinoflagellates in corals. In The 79th annual meeting of the botanical society of Japan (Toki Messe, Niigata Convention Center). (2015)
- Luo, Y.-J., Takeuchi, T., Yamada, L., Endo, K., and Satoh, N. Comprehensive analysis of the calcium phosphate shell proteome of the brachiopod, Lingula anatina. BiominXIII, 13th International symposium on biomineralization. (Parque de las CienciasGranada, Spain) (2015)
- Hamada, M., Kürn, U., Schröder, K., Fraune, S., Satoh, N., and Bosch, T.C.G. MOLECULAR INTERACTION AND GENOME EVOLUTION IN THE GREEN HYDRA AND CHLORELLA SYMBIOSIS. International Workshop Animal evolution: new perspectives from early emerging metazoans. (Evangelische Akademie Tutzing, Germany) (2015)
- Nakashima, K. Intestinal mucosal immunity in Ciona intestinalis. In The 86th Annual Meeting of the Zoological Society of Japan (Toki Messe, Niigata Convention Center, Japan). (2015)
- Khalturin, K., Shinzato, C., Khalturina, M., Fujie, M., Koyanagi, R., Goto, H., and Satoh, N. Life cycle regulation in Cnidaria and the evolution of body plans. The 86th Annual Meeting of the Zoological Society of Japan. (Toki Messe, Niigata Convention Center, Japan) (2015)
- Takeuchi, T., Fujie, M., Nagai, K., and Satoh, N. Development of linkage map of the pearl oyster Pinctada fucata using GGRS (Genotyping by Genome Reducing and Sequencing). In The Japanese Society of Fisheries Science (Tohoku University). (2015)
- Zayasu, Y. Comparison of molecular phylogenetic relationships between massive corals and pit crabs. In International Symposium: Phylogenetic, ecological, and behavioural aspects among cryptic or semi-cryptic species of Crustacea, Carcinological Society of Japan 53rd Annual Meeting (Shinagawa Campus, Tokyo University of Marine Science and Technology). (2015)
- Maeda, K., and Aizawa, M. Butis butis living in Manko. In Okinawa Fish Seminar (Okinawa Prefectural Nago Youth Center). (2015)
- Satoh, N. The dorsal midline hypothesis for chordate origin. Academia Sinica – OIST Joint Meeting“Evolution of Complex Systems”. (Institute of Cellular and Organismic Biology, Academia Sinica, Taipei, Taiwan) (2015)
- Arimoto, A., Nishitsuji, K., Shinzato, C., Satoh, N., and Shoguchi, E. The genome assembly of the macro green alga Caulerpa lentillifera. Academia Sinica - OIST Joint Meeting “Evolution of Complex Systems”. (Institute of Cellular and Organismic Biology, Academia Sinica, Taipei, Taiwan) (2015)
- Baughman, K., Hisata, K., Shoguchi, E., and Satoh, N. Genome analysis of the corallivorous starfish Acanthaster planci. Academia Sinica - OIST Joint Meeting “Evolution of Complex Systems”. (Institute of Cellular and Organismic Biology, Academia Sinica, Taipei, Taiwan) (2015)
- Hamada, M., Kürn, U., Schröder, K., Fraune, S., Khalturina, M., Shinzato, C., Bosch, T.C.G., and Satoh, N. Molecular interaction and genome evolution in the green hydra and Chlorella symbiosis. Academia Sinica - OIST Joint Meeting “Evolution of Complex Systems”. (Institute of Cellular and Organismic Biology, Academia Sinica, Taipei, Taiwan) (2015)
- Ikegami, K., Hamada, M., and Satoh, N. Toward an understanding of the phylotypic stage in the ascidian Ciona intestinalis. Academia Sinica - OIST Joint Meeting “Evolution of Complex Systems”. (Institute of Cellular and Organismic Biology, Academia Sinica, Taipei, Taiwan) (2015)
- Khalturin, K., Shinzato, C., Khalturina, M., Fujie, M., Koyanagi, R., Goto, H., and Satoh, N. Evolution of body plans and life cycle regulation in Cnidaria. Academia Sinica - OIST Joint Meeting “Evolution of Complex Systems”. (Institute of Cellular and Organismic Biology, Academia Sinica, Taipei, Taiwan) (2015)
- Lu, T.-M., Furuta, H., and Satoh, N. Phylogenomic study of Dicyema japonicum reveals the evolutionary position of the phylum Dicyemida within Spiralia. Academia Sinica - OIST Joint Meeting “Evolution of Complex Systems”. (Institute of Cellular and Organismic Biology, Academia Sinica, Taipei, Taiwan) (2015)
- Luo, Y.-J., and Satoh, N. The Lingula genome and the patterning of BMP-chordin axis in brachiopods. Academia Sinica - OIST Joint Meeting “Evolution of Complex Systems”. (Institute of Cellular and Organismic Biology, Academia Sinica, Taipei, Taiwan) (2015)
- Nakashima, K., Kimura, S., Ogawa, Y., Wada, M., Yamada, L., Sawada, H., Kikuchi, S., and Satoh, N. Chitin-based barrier immunity in the gut of the ascidian tunicate Ciona intestinalis. Academia Sinica - OIST Joint Meeting “Evolution of Complex Systems”. (Institute of Cellular and Organismic Biology, Academia Sinica, Taipei, Taiwan) (2015)
- Nishitsuji, K., Arimoto, A., Fujie, M., Shinzato, C., and Satoh, N. The genome project of the Okinawan brown algae, Mozuku, Cladosiphon okamuranus and Nemacystus decipiens. Academia Sinica - OIST Joint Meeting “Evolution of Complex Systems”. (Institute of Cellular and Organismic Biology, Academia Sinica, Taipei, Taiwan) (2015)
- Yasuoka, Y., Koyanagi, R., Yasuoka, Y., Shinzato, C., and Satoh, N. The “mesodermal” gene brachyury regulates ectoderm-endoderm demarcation in a diploblastic animal, the scleractinian coral Acropora digitifera. Academia Sinica – OIST Joint Meeting“Evolution of Complex Systems”. (Institute of Cellular and Organismic Biology, Academia Sinica, Taipei, Taiwan) (2015)
- Shinzato, C., Mungpakdee, S., Arakaki, N., and Satoh, N. Genome-wide SNP analysis explains coral diversity and recovery in the Ryukyu Archipelago. Academia Sinica - OIST Joint Meeting “Evolution of Complex Systems”. (Institute of Cellular and Organismic Biology, Academia Sinica, Taipei, Taiwan) (2015)
- Shoguchi, E., Shinzato, C., Hisata, K., and Satoh, N. Comparative genomics of symbiotic marine dinoflagellates. Academia Sinica - OIST Joint Meeting “Evolution of Complex Systems”. (Institute of Cellular and Organismic Biology, Academia Sinica, Taipei, Taiwan) (2015)
- Zayasu, Y., Nakajima, Y., Nishitsuji, Y., Yasuoka, Y., Sakai, K., Suzuki, G., Satoh, N., and Shinzato, C. Population structure and genetic diversity among Acropora tenuis (Scleractinia, Cnidaria) in Nansei Islands, Okinawa, Japan. Academia Sinica - OIST Joint Meeting “Evolution of Complex Systems”. (Institute of Cellular and Organismic Biology, Academia Sinica, Taipei, Taiwan) (2015)
- Zayasu, Y. The benefit from POGO training program, current research interests and involvement with NANO. POGO-17 (Partnership for Observation of the Global Oceans 17th Annual Meeting). (Minatomirai 21, Queen Mall Conference room, Yokohama, Japan) (2016)
- Arimoto, A., Nishitsuji, K., Shinzato, C., Shoguchi, E., and Satoh, N. Genomic and metagenomic analysis of Caulerpa lentillifera. In The 40th Meeting of Japanese Society of Phycology (Nippon Dental Univerisity, Tokyo). (2016)
- Nishitsuji, K., Arimoto, A., Fujie, M., Arakaki, N., Shinzato, C., Shoguchi, E., and Satoh, N. Genome analysis of Phaeophyceae Chodariales, Cladosiphon okamuranus. In The 40th Meeting of Japanese Society of Phycology (Nippon Dental Univerisity, Tokyo). (2016)
- Shoguchi, E., Shinzato, C., Hisata, K., and Satoh, N. Three Symbiodinium genomes and the diversity. In The 40th Meeting of Japanese Society of Phycology (Nippon Dental Univerisity, Tokyo). (2016)
- Zayasu, Y., Nakajima, Y., Sakai, K., Suzuki, G., Satoh, N., and Shinzato, C. Determining source and sink populations of corals in the Nansei Islands, Japan. Island Environment and Climate Change in Asia-Pacific Region. (OIST Seaside House) (2016)
- Arimoto, A., Nishitsuji, K., Higa, Y., Shinzato, C., Shoguchi, E., and Satoh, N. Genomic analysis of Caulerpa lentillifera for the efficient cultivation and breeding. In The Japanese Society of Fisheries Science Spring Meeting 2016 (Tokyo University of Marine Science and Technology). (2016)
- Nishitsuji, K., Arimoto, A., Iwai, K., Fujie, M., Arakaki, N., Shinzato, C., Satoh, N., and Shoguchi, E. The Brown Alga, Cladsiphon okamuranus: from genome to biosynthesis. In The Japanese Society of Fisheries Science Spring Meeting 2016 (Tokyo University of Marine Science and Technology). (2016)
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 OIST Marine Genomics Seminar
- Title: Genus Vigna -the Wild and Sexy-
- Speaker: Dr. Ken Naito (Genetic Resource Center, National Institute of Agrobiological Sciences)
- Date: Nov 20 (Friday) 13:00 to 14:00
- Venue : OIST Lab1 Level-C Meeting Room (C016)
6.2 OIST Marine Genomics Seminar
- Title: Enhancer sequence to tissue specificity during development
- Speaker: Dr. Emma Farley (University of California, Berkeley, USA)
- Date: July 24 (Friday) 10:00 to 11:00
- Venue : OIST Lab1 Level-C Meeting Room (C016)
6.3 OIST Marine Genomics Seminar
- Title: Corals under stress - A study of the coral immune system
- Speaker: Dr. Jeroen van de Water (James Cook University)
- Date: 13:00 - 14:00 April 13, 2015
- Venue: OIST Lab1 Level-D Meeting Room (D015), Okinawa, Japan
7. Other
Nothing to report.



