FY2013 Annual Report
Marine Genomics Unit
Professor Nori Satoh

Abstract
The genome contains all the genetic information of a given organism. Decoding the genome therefore provides the molecular basis for understanding every biological phenomenon. Since the turn of the 21st century, genomes of various metazoans have been sequenced, and consequently studies progressed efficiently in the fields of evolutionary biology, developmental biology, and environmental biology. The objective of the Marine Genomic Unit (MGU) is to decode the genomes of target marine invertebrates so as to comprehensively elucidate the molecular mechanisms underlying the (a) environmental responses, (b) development and evolution of marine invertebrates, and (c) specific functions of metazoans. In 2011, we have decoded genome of the coral Acroporal digitifera. The best achievement in fiscal year 2013 was decoding the genome of a zooxanthella (coral-symbiotic brown algae), Symbiodinium minutum. OIST-MGU is therefore a lab in which the genomes of both coral and zooxanthella have been sequenced.
(a) Environmental genomics:
The coral reefs of the Okinawa islands are amongst the most biologically diverse ecosystems in the world. The key organisms in their establishment, the scleractinian corals, increasingly face a range of human-caused challenges including seawater temperature rises and ocean acidification. To understand better the molecular mechanisms underlying coral biology, we succeeded in deciphering the 420-Mbp-long genome of the coral Acropora digitifera in 2011, and found that the coral innate immunity repertoire is notably more complex than that of the sea anemone. This year, we succeeded in deciphering the 1500-Mbp-long genome of a zooxanthella, Symbiodinium minutum. We are now conducting experiments, taking advantages of the genomic information to elucidate molecular mechanisms underlying the establishment and collapse of biosymbiosis between corals and zooxanthella.
(b) Developmental and evolutionary genomics:
We are interested in the origin and evolution of chordates. We have already analyzed an evolutionary scenario within three groups of chordates. Since chordates originated from common ancestor shared with non-chordate deuterostomes such as echinoderms and hemichordates, a big question remained is an evolutionary link between non-chordate deuterostomes and chordates. This year, Nori Satoh has published a book titled “Developmental Genomics of Ascidians”. The simplicity and lack of redundancy in their regulatory genes have made ascidians one of the most useful species in studying developmental genomics. In this book, he explains the developmental genomics of ascidians, stresses the simplicity of Ciona developmental system, and emphasizes single-cell level analyses. This book actively accentuates the advantages of using ascidians as model organisms in an up-and-coming field of developmental genomics.
(c) Functional genomics:
The pearl oyster Pinctada fucata is able to produce pearls, which are of significant value both industrially and as jewels. In 2012, we have deciphered the 1500-Mbp-long genome of P. fucata, which is estimated to contain approximately 23,000 protein-coding genes. This year (2013), in collaboration with many researchers in the pearl oyster biology, we have annotated genes involved in various biological processes of this mollusk, including those involved in biomineralization, reproduction, embryological development, and others. These reports have been published in a special issue of Zoological Science, this year.
1. Staff
- Group Leader
- Dr. Takeshi Kawashima
- Dr. Eiichi Shoguchi
- Dr. Chuya Shinzato
- Researcher
- Dr. Mayuko Hamada
- Dr. Ken Maeda
- Dr. Keisuke Nakashima
- Dr. Sutada Mungpakdee
- Dr. Takeshi Takeuchi
- Dr. Fuki Gyoja
- Dr. Yuuri Yasuoka (JSPS, PD Fellow)
- Dr. Konstantin Khalturin
- Lab rotation students
- Kenneth Baughman
- Keita Ikegami
- Yi-Jyun Luo
- Technical Staff
- Ms. Kanako Hisata
- Ms. Sakura Kikuchi
- Dr. Makiko Tanaka
- Dr. Miyuki Kanda
- Dr. Mariia Khalturina
- Research Assistant
- Yuki Yasuoka
- Saori Araki
- Research Administrator
- Ms. Shoko Yamakawa
- Ms. Tomomi Teruya
2. Collaborations
- Theme: Decoding of the Symbiodinium genomes
- Type of collaboration: Collaboration
- Researchers:
- Dr. Mónica Medina, Penn State University, USA
- Dr. , State University of New York, USA
- Theme: Chip-sequence analysis of Ci-Bra target genes
- Type of collaboration: Collaboration
- Researchers:
- Dr. Hiroki Takahashi, National Institute for Basic Biology, Japan
- Theme: Analysis of Ciona genes with transgenic lines
- Type of collaboration: Collaboration
- Researchers:
- Dr. Yasunori Sasakura, Shimoda Marine Research Center, University of Tsukuba, Japan
- There are several collaborative works not specifically listed here.
3. Activities and Findings
3.1 Environmental genomics
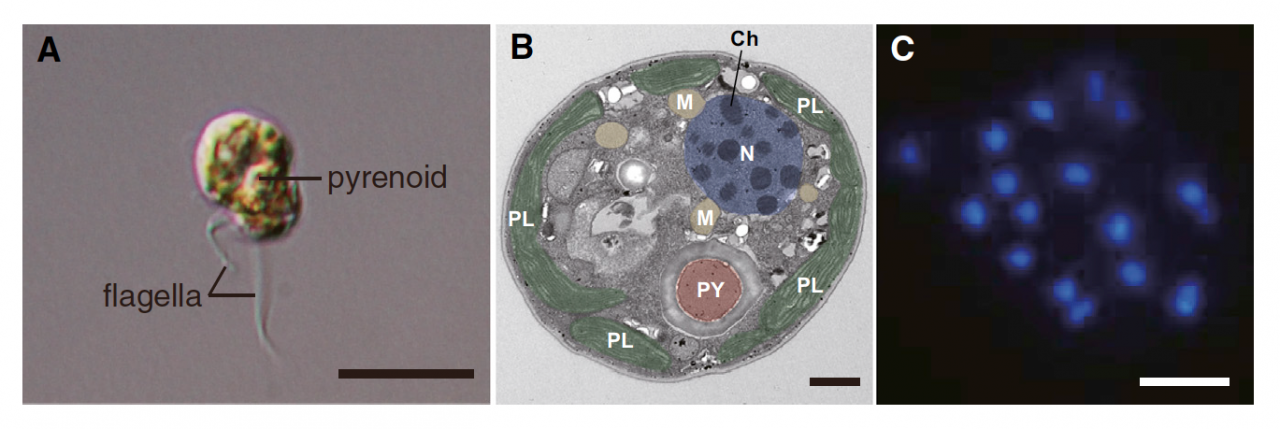
To understand the coral biology, deciphering genomes of both corals and their symbionts (Symbiodinium) is essential. In 2011, we succeeded in deciphering the 420-Mbp-long genome of the coral Acropora digitifera. Therefore, decoding of the Symbiodinium genome has especially been desired. Symbiodinium are a member of Dinoflagellata, which are ecologically and economically important, unicellular eukaryotes that inhabit both marine and freshwater habitats. Most dinoflagellates are 10–100 mm in diameter and are characterized by two flagella, with a unique cell covering referred to as the theca (Figure 1). They show remarkable characteristics such as permanently condensed, liquid-crystalline chromosomes (Figure 1). However, dinoflagellates possess some of the largest nuclear genomes known among eukaryotes (1,500–245,000 Mbp), which have previously thwarted whole-genome sequencing of members of this lineage. We challenged this project by selecting a culturable dinoflagellate, Symbiodinium minutum (Figure 1), which has one of the smallest reported dinoflagellate genomes (1,500 Mbp).
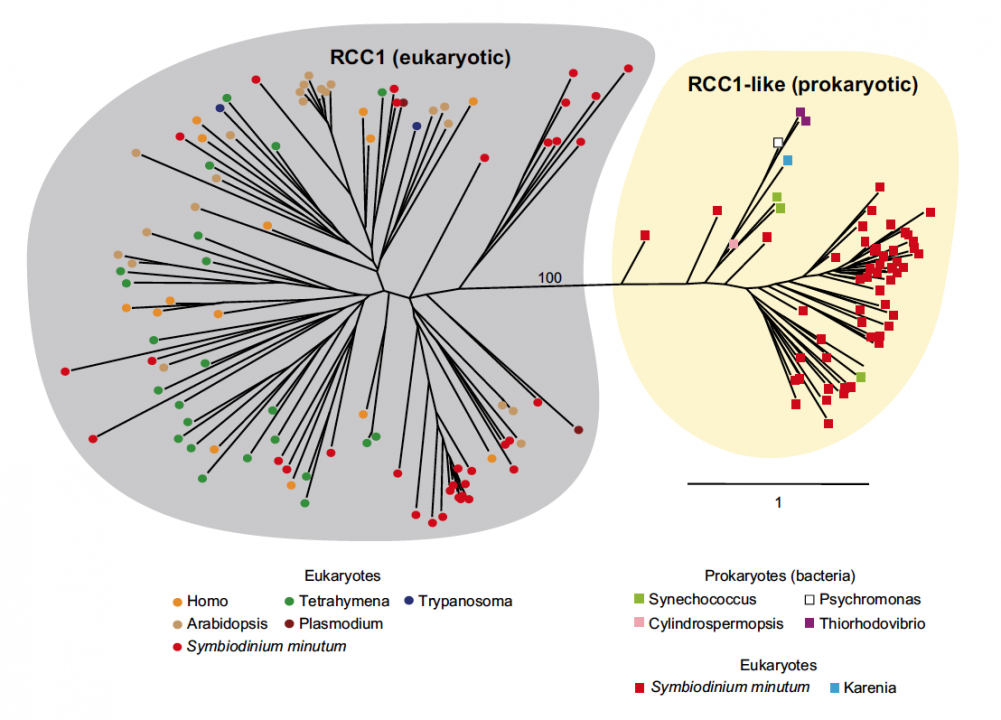
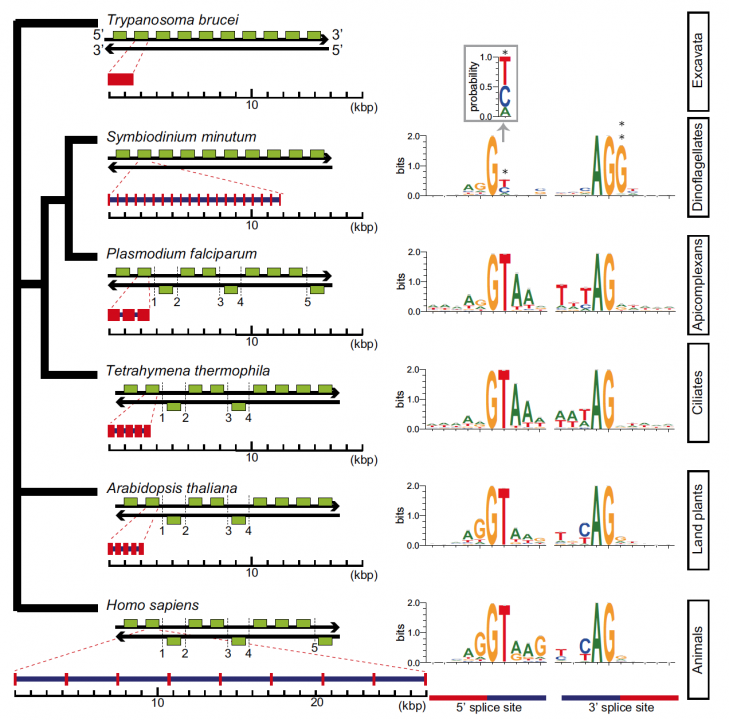
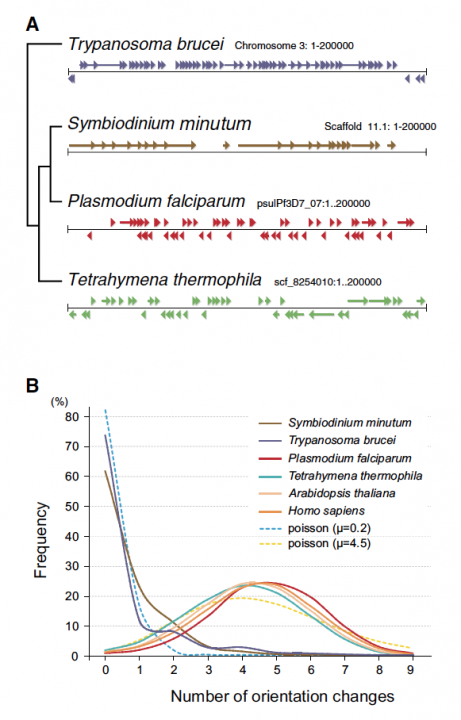
Sequencing reads from S. minutum were assembled into 616 Mbp gene-rich DNA regions that represented roughly half of the estimated 1,500 Mbp genome of this species. The assembly however encoded approximately 42,000 protein-coding genes, consistent with previous dinoflagellate gene number estimates using transcriptomic data. The Symbiodinium genome contains duplicated genes for regulator of chromosome condensation proteins, nearly one-third of which have eukaryotic orthologs, whereas the remainder have most likely been acquired through bacterial horizontal gene transfers (Figure 2). Symbiodinium genes are enriched in spliceosomal introns (mean =18.6 introns/gene) (Figure 3). Donor and acceptor splice sites are unique, with 50 sites utilizing not only GT but also GC and GA, whereas at 30 sites, a conserved G is present after AG (Figure 3). All spliceosomal snRNA (U1–U6) are clustered in the genome. Surprisingly, the Symbiodinium genome displays unidirectionally aligned genes throughout the genome, forming a cluster-like gene arrangement (Figure 4). Such alignment of genes is only known in Trypanosoma genomes. Our data elucidate the organization and gene inventory of dinoflagellates and lay the foundation for future studies of this remarkable group of eukaryotes. The genomic information is also fundamental in research of coral-symbiont symbiosis.
3.2 Developmental and evolutionary genomics:
We are interested in the origin and evolution of chordates. We have already analyzed an evolutionary scenario within three groups of chordates. Since chordates originated from common ancestor shared with non-chordate deuterostomes such as echinoderms and hemichordates, a big question remained is an evolutionary link between non-chordate deuterostomes and chordates. This year, Nori Satoh has published a book titled “Developmental Genomics of Ascidians”. The simplicity and lack of redundancy in their regulatory genes have made ascidians one of the most useful species in studying developmental genomics. In this book, he explains the developmental genomics of ascidians, stresses the simplicity of Ciona developmental system, and emphasizes single-cell level analyses. This book actively accentuates the advantages of using ascidians as model organisms in an up-and-coming field of developmental genomics.
In addition, this year, we have identified an intact ParaHox cluster with temporal colinarity but altered spatial colinarity in the hemichordate Ptychodera flava. Since we reported last year that hemichordates P. flava and Saccoglossus kowalevskii contain an intact Hox cluster within approximately 500-kbp-long genomic region of each species, this indicates similarity of genomic organization between non-chordate acorn worms and amphioxus, a most basal chordate.
3.3 Functional genomics:
The pearl oyster Pinctada fucata is able to produce pearls, which are of significant value both industrially and as jewels. In 2012, we have deciphered the 1500-Mbp-long genome of P. fucata, which is estimated to contain approximately 23,000 protein-coding genes. This year (2013), in collaboration with many researchers in the pearl oyster biology, we have annotated genes involved in various biological processes of this mollusk, including those involved in biomineralization, reproduction, embryological development, and others. These reports have been published in a special issue of Zoological Science, this year.
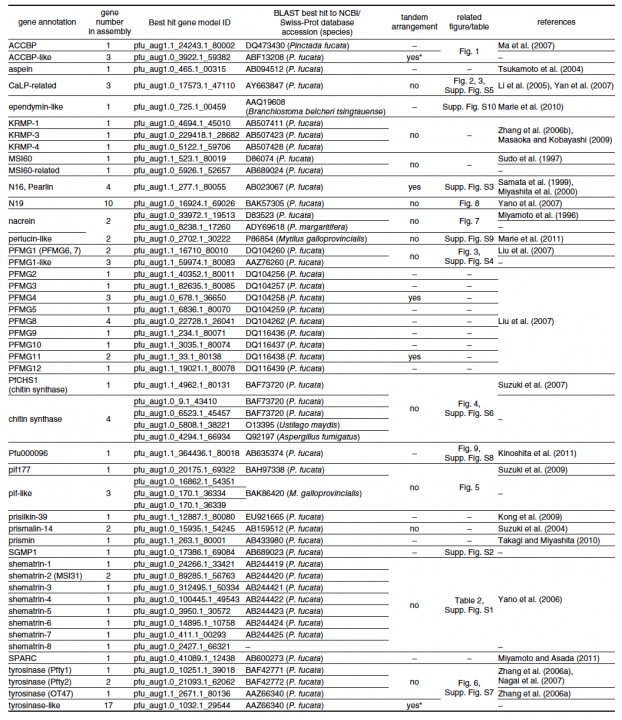
For example, in molluscs, shell matrix proteins are associated with biomineralization, a biologically controlled process that involves nucleation and growth of calcium carbonate crystals. Identification and characterization of shell matrix proteins are important for better understanding of the adaptive radiation of a large variety of molluscs. We searched the draft genome sequence of the pearl oyster and annotated 30 different kinds of shell matrix proteins (Table 1). Of these, we could identify Perlucin, ependymin-related protein and SPARC as common genes shared by bivalves and gastropods; however, most gastropod shell matrix proteins were not found in the P. fucata genome. Glycinerich proteins were conserved in the genus Pinctada. Another important finding with regard to these annotated genes was that numerous shell matrix proteins are encoded by more than one gene; e.g., three ACCBP-like proteins, three CaLPs, five chitin synthase-like proteins, two N16 proteins (pearlins), 10 N19 proteins, two nacreins, four Pifs, nine shematrins, two prismalin-14 proteins, and 21 tyrosinases. This diversity of shell matrix proteins may be implicated in the morphological diversity of mollusc shells. The annotated genes reported here can be searched in P. fucata gene models version 1.1 and genome assembly version 1.0 (http://marinegenomics.oist.jp/pinctada_fucata). These genes should provide a useful resource for studies of the genetic basis of biomineralization and evaluation of the role of shell matrix proteins as an evolutionary toolkit among the molluscs.
4. Publications
4.1 Journals
(a) Environmental genomics:
Shoguchi1, E., Shinzato, C., Kawashima, T., Gyoja, F., Mungpakdee, S., Koyanagi, R., Takeuchi, T., Hisata, K., Tanaka, M., Fujiwara, M., Hamada, M., Azadeh, S., Fujie, M., Usami, T., Goto, H., Yamasaki, S., Arakaki, N., Suzuki, Y., Sugano, S., Toyoda, A., Kuroki, Y., Fujiyama, A., Medina, M., Coffroth, M. A., Bhattacharya, D., and Satoh, N. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Current Biology 23, 1399-1408. (2013)
Husnik, F., Nikoh, N., Koga, R., Ross, L., Duncan, R. P., Fujie, M., Tanaka, M., Satoh, N., Bachtrog, D., Wilson, A. C. C., Dohlen, C. D., Fukatsu, T., and McCutcheon, J. P. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 153, 1567–1578. (2013)
Sota, T., Konuma, J., Fujiwara, M., Shoguchi, E. Genome sizes of three species in the subtribe Carabina (Coleoptera: Carabidae) Entomological Science 16: 122-124. (2013)
Shinzato C, Inoue M, Kusakabe M. A snapshot of a coral “holobiont”: A transcriptome assembly of the scleractinian coral, Porites, captures a wide variety of genes from both the host and symbiotic zooxanthellae. PLoS ONE 9(1): e85182. (2014)
Tsuta H, Shinzato C, Satoh N, Hidaka M. Telomere Shortening in the Colonial Coral Acropora digitifera During Development. Zoolog Sci 31(3):129-34. (2014)
Shoguchi, E., Tanaka, M., Takeuchi, T., Shinzato, C., Satoh, N. Probing a coral genome for components of the photo-protective scytonemin biosynthetic pathway and the 2-aminoethylphosphonate pathway. Marine Drugs, 11(2):559-70. (2013)
(b) Developmental and evolutionary genomics:
Ikuta, T., Chen, Y.C., Annunziata, R., Ting, H.C., Tung, C., Koyanagi, R., Tagawa, K., Humphreys, T., Fujiyama, A., Saiga, H., Satoh, N., Yu, J. K., Arnone, M.I., Su, Y.H. Identification of an intact ParaHox cluster with temporal colinearity but altered spatial colinearity in the hemichordate Ptychodera flava. BMC Evolutionary Biology 13:129. (2013)
Kobayashi, K., Yamada, L., Satou, Y., Satoh, N. Differential gene expression in notochord and nerve cord fate segregation in the Ciona intestinalis embryo. Genesis 51:647–659. (2013)
Matsumae ,H., Hamada, M., Fujie, M., Niimura, Y., Tanaka, H., Kawashima, T. A methodical microarray design enables surveying of expression of a broader range of genes in Ciona intestinalis. Gene 519: 82-90. (2013)
Satoh, N. Ciona: a Model for Developmental Genomics. Encyclopedia of Life Sciences on line. (2013)
GIGA Community of Scientists, Bracken-Grissom H, Collins AG, Collins T, Crandall K, Distel D, Dunn C, Giribet G, Haddock S, Knowlton N, Martindale M, Medina M, Messing C, O'Brien SJ, Paulay G, Putnam N, Ravasi T, Rouse GW, Ryan JF, Schulze A, Wörheide G, Adamska M, Bailly X, Breinholt J, Browne WE, Diaz MC, Evans N, Flot JF, Fogarty N, Johnston M, Kamel B, Kawahara AY, Laberge T, Lavrov D, Michonneau F, Moroz LL, Oakley T, Osborne K, Pomponi SA, Rhodes A, Santos SR, Satoh N, Thacker RW, Van de Peer Y, Voolstra CR, Welch DM, Winston J, Zhou X. The Global Invertebrate Genomics Alliance (GIGA): developing community resources to study diverse invertebrate genomes. J Hered 105(1):1-18. (2014)
Fuchs B, Wang W, Graspeuntner S, Li Y, Insua S, Herbst EM, Dirksen P, Böhm AM, Hemmrich G, Sommer F, Domazet-Lošo T, Klostermeier UC, Anton-Erxleben F, Rosenstiel P, Bosch TC, Khalturin K. Regulation of polyp-to-jellyfish transition in Aurelia aurita. Curr Biol. 24:263-73. (2014)
(c) Functional genomics:
Takeuchi, T., Kawashima, T., Koyanagi, R., Satoh, N. The genome of the Japanese pearl oyster, Pinctada fucata. In "Recent Advances in Pearl Research".Eds. S. Watabe, K. Maeyama, H. Nagasawa, TERRAPUB, Tokyo (2013)
Kawashima, T., Takeuchi, T., Koyanagi, R., Kinoshita, S., Endo, H., Endo, K. Initiating the Mollusk Genomics Annotation Community: Toward Creating the Complete Curated Gene-Set of the Japanese Pearl Oyster, Pinctada fucata. Zoological Science 30: 794–796. (2013)
Koyanagi, R., Takeuchi, T., Hisata, K., Gyoja, F., Satoh, N., Kawashima, T. MarinegenomicsDB: An integrated genome viewer for community-based annotation of genomes. Zoological Science 30: 797–800. (2013)
Miyamoto, H., Endo, H., Hashimoto, N., Iimura, K., Isowa, Y., Kinoshita, S., Kotaki, R., Masaoka, T., Miki, T., Nakayama, S., Nogawa, C., Notozawa, A., Ohmori, F., Sarashina, I., Suzuki, M., Takagi, R., Takahashi, J., Takeuchi, T., Yokoo, N., Satoh, N., Toyohara, H., Miyashita, T., Wada, H., Samata, T., Endo, K., Nagasawa, H., Asakawa, S., Watabe, S. The diversity of shell matrix proteins: genome-wide investigation of the pearl oyster Pinctada fucata. Zoological Science 30: 801–816. (2013)
Morino, Y., Okada, K., Niikura, M., Honda, M., Satoh, N., Wada, H. A genome-wide survey of genes encoding transcription factors in the Japanese pearl oyster, Pinctada fucata: I. Homeobox genes. Zoological Science 30: 851–857. (2013)
Matsumoto, T., Masaoka, T., Fujiwara, Y., Nakamura, Y., Kinoshita, S., Satoh, N., Awaji, M. Reproduction-related genes in the pearl oyster genome. Zoological Science 30: 826–850. (2013)
Koga, H., Hashimoto, N., G. Suzuki, D., Ono, H., Yoshimura, M., Suguro, T., Yonehara, Y., Abe, T., Satoh, N., Wada, H. A genome-wide survey of genes encoding transcription factors in Japanese pearl oyster Pinctada fucata II. Tbx, Fox, Ets, HMG, NFκB, bZIP and C2H2 Zinc fingers. Zoological Science 30: 858–867. (2013)
Gyoja, F., Satoh, N. Evolutionary aspects of variability in bHLH orthologous families: insights from the pearl oyster Pinctada fucata. Zoological Science 30: 868–876. (2013)
(d) Other Fields:
Kondo, M., Maeda, K., Hirashima, K., Tachihara, K. Comparative larval development of three amphidromous Rhinogobius species making reference to their habitat preferences and migration biology. Marine & Freshwater Research 64:249-466. (2013)
Maeda K., Tan H.H. Review of Stiphodon (Gobiidae: Sicydiinae) from western Sumatra, with description of a new species. Raffles Bulletin of Zoology 62:749-761. (2013)
Maeda, K., Saeki, T. First record of a sicydiine goby, Stiphodon multisquamus (Actinopterygii: Gobioidei: Gobiidae), from Okinawa Island, Japan. Species Diversity 18:215–221. (2013)
Nishi,S., Tsubouchi, T., Takaki, Y., Koyanagi, R., Satoh, N., Maruyama, T., Hatada, Y. Draft genome sequence of Loktanella cinnabarina LL-001T, isolated from deep-sea floor sediment. Genome Announcements, 1(6) doi: 10.1128/genomeA.00927-13. (2013)
4.2 Books and other one-time publications
(a) Environmental genomics:
Satoh N. Developmental Genomics of Ascidians. Wiley Brackwell, New York. (2014)
4.3 Oral and Poster Presentations
[Oral presentations]
Maeda, K. & Tachihara, K. Larval study to find crypto-benthic gobies, in 9th Indo-Pacific Fish Conference, Okinawa Convention Center, Okinawa, Japan (2013.6.26).
Satoh, N. Cross talk of coral and Symbiodinium genes during environmental changes, in Gordon Research Conference “Marine Molecular Ecology”, The Hong Kong University of Science and Technology, Hong Kong, China (2013.8.13).
Takeuchi, T., Kawashima, T., Koyanagi, R., Endo, K. & Satoh, N. Genome-wide survey for biomineralization genes in the Japanese pearl oyster, Pinctada fucata, in The 12th International Symposium on Biomineralization, Friberg, Saxony, Germany (2013.8.30).
Hamada, M., KÜRN, U., SCHRÖDER, K., FRAUNE, S., Satoh, N. & BOSCH, TCG. The Hydra viridissima / Chlorella symbiosis revisited: understanding a complex team of players at the base of animal evolution, in International Workshop "Unravelling the Developmental Regulatory Network in Early Animals", Evangelische Akademie Tutzing, Germany (2013.9.25).
Shinzato, C. Biological traits of corals revealed by the genome project, in Oregon State University Integrative Biology Winter 2014 Seminar Series, Oregon State University, USA (2014.2.10).
Shoguchi, E. A first assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure, in Oregon State University Integrative Biology Winter 2014 Seminar Series, Oregon State University, USA (2014.2.10).
[Poster presentation]
Luo, YJ., Koyanagi, R., Tanaka, M., Endo, K. & Satoh, N. Decoding the genome of brachiopod Lingula anatina and exploring the patterning mechanisms of its body plan, in EMBO Practical Course: Marine animal models in evolution & development, Fiskebäckskil, Sweden (2013.7.1).
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 OIST Marine Genomics Seminar
- Title: Chemistry and Biology of Okinawan Marine natural products.
- Speaker: Dr. Junichi Kobayashi (Hokkaido University, Japan)
- Date: 16:00 – 17:00 Jun 20, 2013
- Venue: OIST Lab1 Level-C Meeting Room 1 (C016), Okinawa, Japan
6.2 OIST Marine Genomics Seminar
- Title: Deciphering basic signaling architectures patterning animal body axes.
- Speaker: Dr. Hiroshi Watanabe (Centre for Organismal Studies (COS), University of Heidelberg)
- Date: 13:00 - 14:00 Dec 10, 2013
- Venue: OIST Lab1 Level-C Meeting Room 1 (C016), Okinawa, Japan
6.3 OIST Marine Genomics Seminar
- Title: Reproductive Strategies Shared by Animals and Plants: Ascidian Self-incompatibility System and Cnidarian Gamete Fusion.
- Speaker: Dr. Hitoshi Sawada (Sugashima Marine Biological Laboratory, Graduate School of Science, Nagoya University)
- Date: 11:00 - 12:00 Jan 22, 2014
- Venue: OIST Lab1 Level-C Meeting Room (C015), Okinawa, Japan
6.4 OIST Marine Genomics Seminar
- Title: Coral disease across the Indo-Pacific
- Speaker: Dr. Greta Smith Aeby (Hawaii Institute of Marine Biology)
- Date: 16:00 - 17:00 March 13, 2014
- Venue: OIST Lab1 Level-D Meeting Room (D015), Okinawa, Japan
6.5 OIST Winter Course "Evolution of Complex Systems" (OWECS) 2013
- Date: 2-7 December, 2013
- Venue: OIST Seaside House, Okinawa, Japan
- Organizers
- Nori Satoh (OIST, Japan)
- Dr. Michael Levine, University of California, Berkeley, USA
- Speakers:
- Michael Levine (University of California, Berkeley, USA)
- Robb Krumlauf (Stowers Institute for Medical Research, USA)
- Nipam H. Patel (University of California, Berkeley, USA)
- Daniel S. Rokhsar (University of California, Berkeley, USA)
- Nori Satoh (OIST, Japan)
- J. K. Sky Yu (Academia Sinica, Taiwan)
- Yi-Hsien Su (Academia Sinica, Taiwan)
- Chris Amemiya (Benaroya Research Institute at Virginia Mason, USA)
- Chris Lowe (Hopkins Marine Station, Stanford University, USA)
- Takehiro Kusakabe (Konan University, Japan)
- David Miller (James Cook University, Australia & JSPS Fellow)
- Konstantin Khalturin(OIST, Japan)
- Kohji Hotta (Keio Univeristy, Japan)
- Yi-Jyun Luo (OIST, Japan)
7. Other
Nothing to report.



