FY2018 Annual Report

[2018.6.14]
Abstract
The genome contains all the genetic information of a given organism. Decoding the genome therefore provides the molecular basis for understanding every biological phenomenon. Since 2009, the Marine Genomics Unit (MGU) has conducted research in the realm of genome-based biological sciences. By decoding genomes of target marine organisms (mainly invertebrates), we wish to understand genetic and developmental mechanisms underlying diversity of marine organisms. The major research fields are (a) evolutionary and developmental genomics of marine invertebrates, (b) environmental genomics of coral reefs, and (c) functional genomics of marine organisms, including algae. We have decoded genome of a coral in 2011, a pearl oyster in 2012, and symbiotic dinoflagellate (Symbiodinium) in 2013. We further decoded genomes of hemichordates and a brachiopod in 2015, a brown alga in 2016, and Crown-Thorns-Starfish in 2017 in collaboration with Australian researchers. This year we could decode genomes of a nemertean, phoronid, and two dinoflagellate clades. Also, in collaboration with researches of Kyushu University and Tohoku University, we succeeded in decoding the habu snake genome, and in collaboration with Vietnam researchers, we decoded a striped catfish. Main results of this year research shall be reported below.
1. Staff FY2018
- Professor Noriyuki Satoh
- Group Leaders
- Eiichi Shoguchi
- Staff Scientists
- Keisuke Nakashima
- Konstantin Khalturin
- Ken Maeda
- Takeshi Takeuchi
- Jun Inoue
- Postdoctoral Scholars
- Yuuri Yasuoka
- Koki Nishitsuji
- Yuna Zayasu
- Asuka Arimoto
- Students
- Tsai-Ming Lu (JSPS DC Fellow)
- Girish Beedessee (JSPS DC Fellow)
- Yafei Mao (JSPS DC Fellow)
- Technical Staffs
- Kanako Hisata
- Sakura Kikuchi
- Seiya Kitanobo
- Haruhi Narisoko
- Hiraku Miyagi (Part-time: 2018/10/1-2019/2/28)
- Research Assistants
- Yuki Yasuoka
- Yoshie Nishitsuji (NEDO project)
- Mayuki Suwa (2018/4/1-)
- Research Administrators
- Shoko Yamakawa
- Tomomi Teruya
2. OIST PhD Graduation
We congratulate a student for his OIST PhD Graduation.
 Thesis Title:
Thesis Title:
Comparative genomic studies on Dicyema japonicum: the phylogenetic position of dicyemids and the genomic adaptations to parasitic lifestyle.
August, 2018.
3. Research activities and findings
3.1 Developmental and evolutionary genomics
Of eight publications in the field of Developmental and Evolutionary Genomics this year, we wish to report here results of two studies, one is of decoding Nemertean and phoronid genomes and the other is on the expansion that enabled chordate muscle evolution.
(1) Many questions remain to be answered to understand the evolution of lophotrochozoans, one of largest groups of protostomes that including leeches, snails and other invertebrates. Lophotrochozoans represent a superphylum that is crucial to our understanding of bilaterian evolution. Nemerteans (ribbon worms) and phoronids (horseshoe worms) are thought to be closely related lophotrochozoans. However, given the inconsistency of molecular and morphological data for these groups, their origins and evolutionary relation with others have been unclear. In this year, we (Luo et al. Nature Ecol. Evol. 2: 141-151, 2018) sequenced draft genomes the nemertean Notospermus geniculatus and the phoronid Phoronis australis, together with transcriptomes along the adult bodies. Our genome-based phylogenetic analyses place Nemertea sister to the group containing Phoronida and Brachiopoda (Fig. 1)
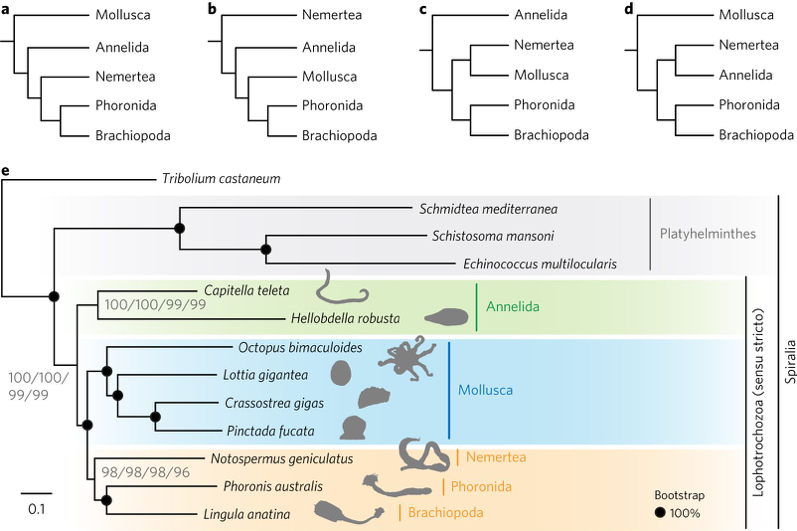
Fig. 1. Genome-based phylogenetics support a close relationship between Nemertea and Phoronida. a–d, Proposed relationships of the five major clades of lophotrochozoans. a, Kryptrochozoa hypothesis (monophyly of Nemertea, Phoronida and Brachiopoda). b, Nemertea as a sister group to other lophotrochozoans. c, Nemertea as a sister group to Mollusca. d, Nemertea as a sister group to Annelida. e, Phylogeny of lophotrochozoans inferred from 173 one-to-one orthologous genes (62,928 amino acid positions with 92% overall matrix completeness). The maximum-likelihood tree was obtained using LG + Γ, LG + I + Γ, LG4M + Γ and LG4X + Γ models with 1,000 bootstrap replicates. Black circles on nodes indicate 100% bootstrap support from all four models.
In addition, we obtained results to show that lophotrochozoans share many gene families with deuterostomes, suggesting that these two groups retain a core bilaterian gene repertoire that ecdysozoans (for example, flies and nematodes) and platyzoans (for example, flatworms and rotifers) do not. Furthermore, an interesting result came from comparative transcriptomics of lophophores. That is, this analysis demonstrates that lophophores of phoronids and brachiopods are similar not only morphologically, but also at the molecular level. Despite dissimilar head structures, lophophores express vertebrate head and neuronal marker genes (Fig. 2). This finding suggests a common origin of bilaterian head patterning, although different heads evolved independently in each lineage. This study reveals a dual nature of lophotrochozoans, where conserved and lineage-specific features shape their evolution.
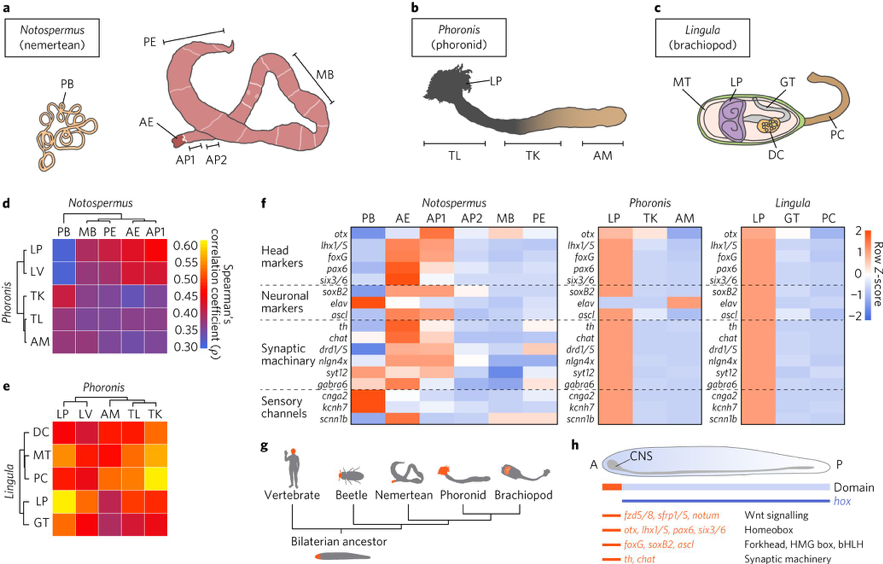
Fig. 2. Comparative transcriptomics reveals molecular similarities between lophophores and bilaterian heads. a–c, Cartoon illustrations of an adult Notospermus (a), Phoronis (b) and Lingula (c; with the dorsal shell removed) with the anterior end facing left. d,e, Analyses of Spearman’s correlation coefficient (ρ) and hierarchical clustering with expression levels of 8,650 orthologous genes from larvae and adult tissues of Notospermus versus Phoronis (d) and Phoronis versus Lingula (e). f, Expression profiles of head patterning-related and neuronal genes in the head of Notospermus and lophophores of Phoronis and Lingula. g,h, Schematic representation of anterior–posterior patterning in bilaterians. g, Simplified phylogeny of bilaterians and regions of anterior positioning heads (highlighted in orange). h, Domain map for conserved signalling components, transcription factors and genes associated with synaptic machinery along the anterior–posterior axis in the last common bilaterian ancestor. A, anterior; AE, anterior end; AM, ampulla; AP1, anterior part 1; AP2, anterior part 2; bHLH, basic helix-loop-helix; CNS, central nervous system; DC, digestive caecum; GT, gut; HMG, high mobility group; LP, lophophore; LV, larva; MB, mid-body; MT, mantle; P, posterior; PB, proboscis; PC, pedicle; PE, posterior end; TK, trunk; TL, trunk with LP.
(2) We are interested in how chordate body plan originated and evolved. We have proposed that fish-like larvae or tadpole-type larvae are foundational to the chordate body plan, given the basal placement of free-living lancelets. That body plan probably made it possible for chordate ancestors to swim by beating a tail formed of notochord and bilateral paraxial muscles. In order to investigate the molecular genetic basis of the origin and evolution of paraxial muscle, we (Inoue and Satoh, Mol. Biol. Evol. 35:914-924, 2018) deduced the evolutionary histories of 16 contractile protein genes from paraxial muscle, based on genomic data from all five deuterostome lineages, using a newly developed orthology identification pipeline and a species tree. As a result, we found that more than twice as many orthologs of paraxial muscle genes are present in chordates, as in non-chordate deuterostomes (ambulacrarians) (Fig. 3). Orthologs of paraxial-type actin and troponin C genes are absent in ambulacrarians and most paraxial muscle protein isoforms diversified via gene duplications that occurred in each chordate lineage. Analyses of genes with known expression sites indicated that some isoforms were reutilized in specific muscles of nonvertebrate chordates via gene duplications. As orthologs of most paraxial muscle genes were present in ambulacrarians, in addition to expression patterns of related genes and functions of the two protein isoforms, regulatory mechanisms of muscle genes are suggested to be considered in future studies of the origin of paraxial muscle.
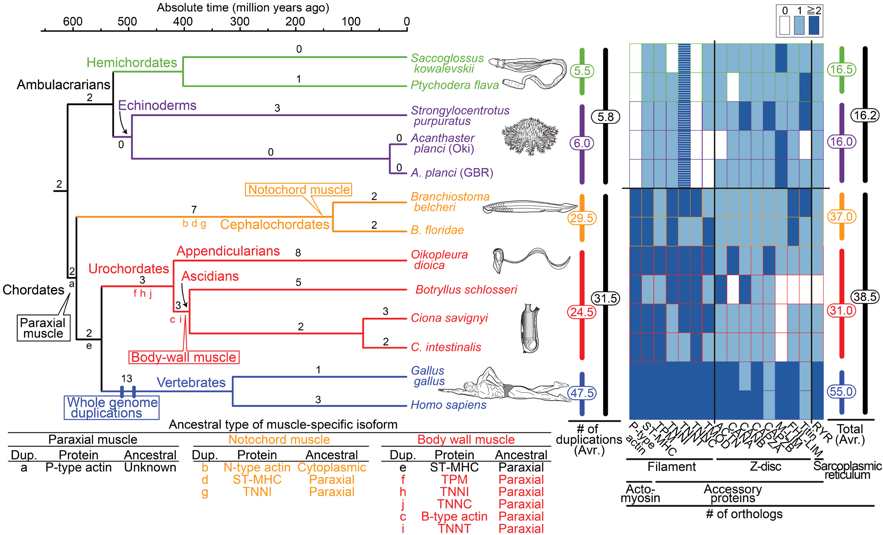
Fig. 3. Time-calibrated deuterostome tree (Holland 1996; Simakov et al. 2015) and evolution of structural (contractile protein) genes of paraxial muscle as a summary of gene tree analyses (supplementary fig. S2A–P, Supplementary Material online)
Numbers at branches are counts of isoform families that experienced gene duplication and numbers at vertical bars (middle) are total counts of gene d uplications in major lineages (supplementary fig. S3, Supplementary Material online). The heat map (right) reflects the estimated number of orthologs (table 1). Striped boxes in ambulacrarian troponin T indicate ambiguity of their functions as in chordate troponin T. The table (below) indicates inferred ancestral types of muscle-specific isoforms.
3.2. Environmental genomics
Marine dinoflagellate, Symbiodinium, is a well-known photosynthetic partner for coral and other diverse, non-photosynthetic hosts in subtropical and tropical shallows, where it comprises an essential component of marine ecosystems. Using molecular phylogenetics, the genus Symbiodinium has been classified into nine major clades, A-I. In 2013, we have decoded first in the world a draft genome of clade B1 Symbiodinium. One of the reported differences among phenotypes is their capacity to synthesize mycosporine-like amino acids (MAAs), which absorb UV radiation. However, the genetic basis for this difference in synthetic capacity is unknown. To understand genetics underlying Symbiodinium diversity, this year we decoded two draft genomes, one from clade A, presumed to have been the earliest branching clade, and the other from clade C, in the terminal branch (Shoguchi et al. BMC Genomics 19:458, 2018).
We obtained results that the nuclear genome of Symbiodinium clade A (SymA) has more gene families than that of clade C, with larger numbers of organelle-related genes, including mitochondrial transcription terminal factor (mTERF) and Rubisco (Fig. 4). While clade C (SymC) has fewer gene families, it displays specific expansions of repeat domain-containing genes, such as leucine-rich repeats (LRRs) and retrovirus-related dUTPases. Interestingly, the SymA genome encodes a gene cluster for MAA biosynthesis, potentially transferred from an endosymbiotic red alga (probably of bacterial origin), while SymC has completely lost these genes. Our analysis demonstrates that SymC appears to have evolved by losing gene families, such as the MAA biosynthesis gene cluster. In contrast to the conservation of genes related to photosynthetic ability, the terminal clade has suffered more gene family losses than other clades, suggesting a possible adaptation to symbiosis. Overall, our results imply that Symbiodinium ecology drives acquisition and loss of gene families.
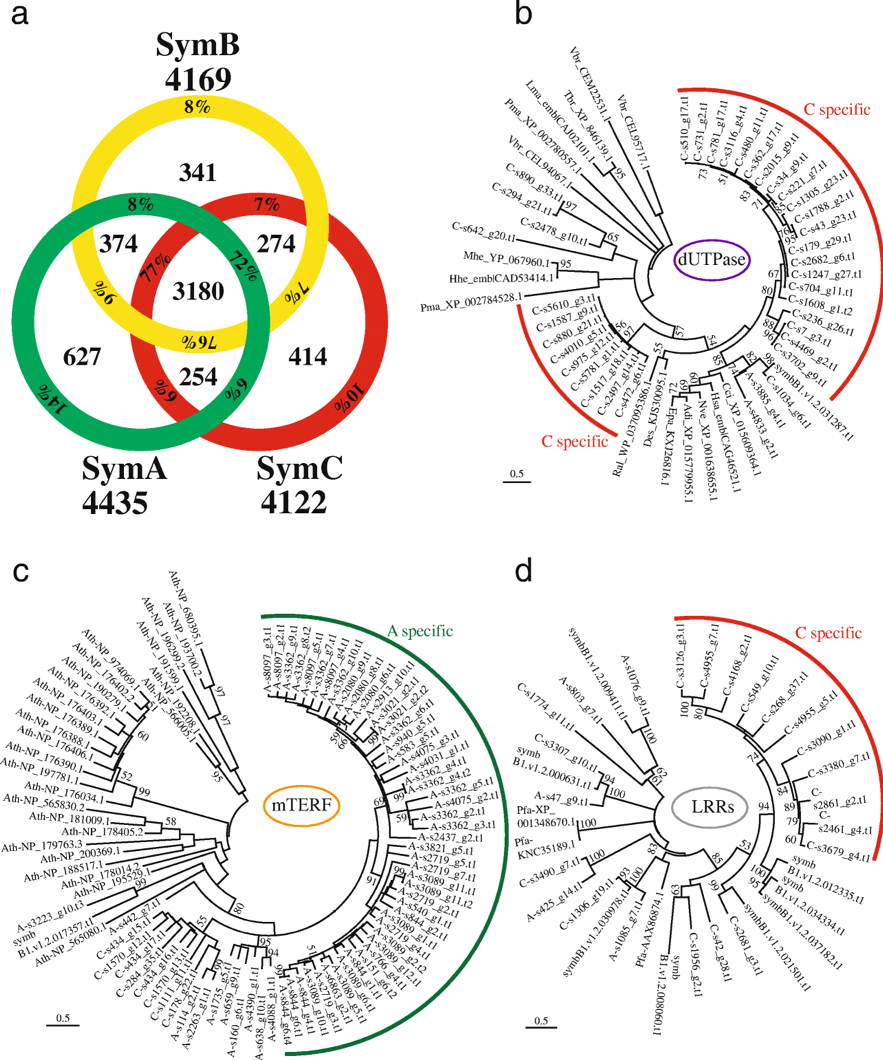
Fig. 4. Comparisons and expansions of gene families in Symbiodinium lineages.
a. Venn diagram comparing Pfam domains among three divergent Symbiodinium genomes. Total numbers of domain types found in each genome are shown outside the circle. Percentages indicate the ratio of the inside number to the total number. b-d. Molecular phylogenies of dUTPase (b), mitochondrial transcription termination (mTERF) proteins (c), and LRR domain-containing proteins (d), respectively. Scale bars show 0.5 changes per site. The arcs show possible lineage specific expansions in SymC (red) and SymA (green), respectively. A, SymA. B, symbB1. C, SymC. Adi, Acropora digitifera. Ath, Arabidopsis thalina. Cci, Cephus cinctus. Des, Desulfatitalea sp. Epa, Exaiptasia pallida. Hhe, Human herpesvirus. Hsa, Homo sapiens. Lma, Leishmania major. Mhe, Macacine herpesvirus. Nve, Nematostella vectensis. Pma, Perkinsus marinus. Ral, Rhizobium alamii. Tbr, Trypanosoma brucei. Vbr, Vitrella brassicaformis
3.3. Functional genomics
Mammalian gut microbiota are integral to host health. However, how this association began remains unclear. This year, we (Nakashima et al., Nature Communications 9: 3402, 2018) show that in basal chordates the gut space is radially compartmentalized into a luminal part where food microbes pass and an almost axenic peripheral part, defined by membranous delamination of the gut epithelium (Fig. 5). While this membrane, framed with chitin nanofibers, structurally resembles invertebrate peritrophic membranes, proteome supports its affinity to mammalian mucus layers, where gut microbiota colonize. In ray-finned fish, intestines harbor indigenous microbes, but chitinous membranes segregate these luminal microbes from the surrounding mucus layer. These data suggest that chitin-based barrier immunity is an ancient system, the loss of which, at least in mammals, provided mucus layers as a novel niche for microbial colonization. These findings provide a missing link for intestinal immune systems in animals, revealing disparate mucosal environment in model organisms and highlighting the loss of a proven system as innovation.
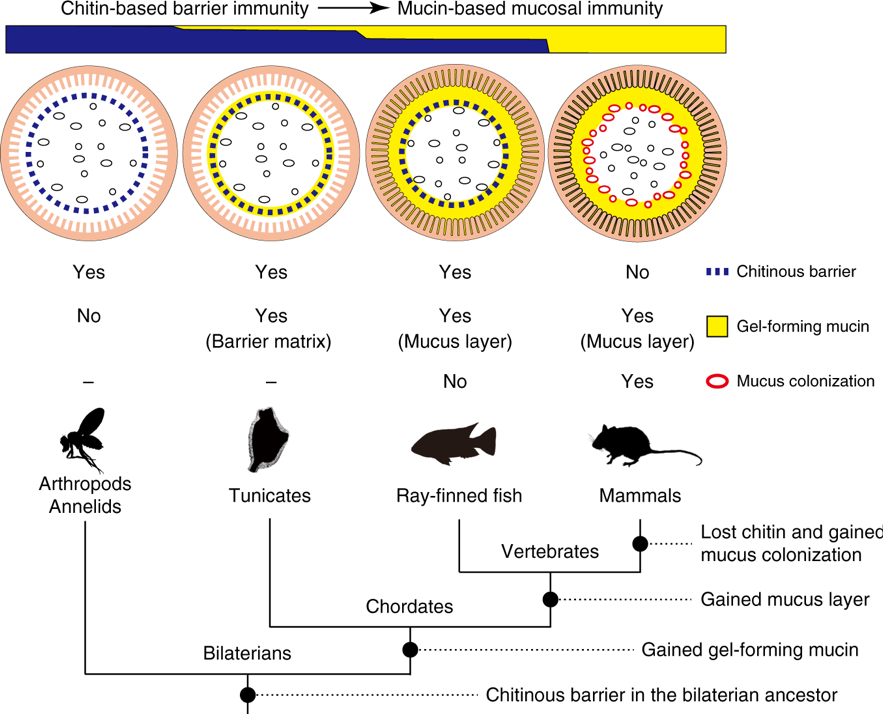
Fig. 5. Transition of gut mucosal surface in chordates and its implication for animal–microbe association. This figure summarizes results of this comparative study of chordates. For animal groups, shown as pictograms, intestinal barrier structures are illustrated above. These illustrations focus on physical, but not cellular or humoral, components of barrier immunity. Arthropods and annelids share chitinous barrier membranes (blue dotted line) that allow movement of nutrients, but not luminal microbes (black ovals), onto the ciliated gut epithelium. This so-called peritrophic matrix (PM) is widely observed in other invertebrates, although the presence of chitin remains unclear. Tunicates possess chitinous membranes embedded in a matrix of gel-forming mucin (yellow circle). This membrane confines food microbes into the luminal space and keeps the ciliated epithelium almost axenic. In ray-finned fish, the mucosal surface is covered with a layer of gel-forming mucin that is secreted from goblet cells. This mucus layer is separated from the luminal, indigenous microbial community by chitinous barrier membranes. In mammals, chitinous membranes no longer exist, and gut microbes directly interact with the surrounding layer of gel-forming mucin. Note that the mammalian mucus system has multiple physiological roles, and there exists regional variation in mucus conditions. This illustration depicts the mouse colon, in which the mucosal surface is covered with two layers of gel-forming mucin, with the outer layer forming a distinct niche for dense microbial colonization (red ovals). “Yes” or “No” indicate the presence or absence of the items listed on the right, respectively. Previously, invertebrate PMs and mammalian mucus layers were not believed to share common descent. New data on tunicates and ray-finned fish, however, fill this gap and suggest a transition from a chitin-based ancestral condition to a mucin-based derived state (top). A tree diagram of animal phylogeny (bottom) helps to infer events that account for the transition (black circles on branches). Mucus colonization in mammalian guts appears to be a novel type of animal–microbe association that was established upon loss of chitin
4. Publications
(a) Developmental and Evolutionary Genomics
- Luo, Y.J., Kanda, M., Koyanagi, R., Hisata, K., Akiyama, T., Sakamoto, H., Sakamoto, T., Satoh, N. Nemertean and phoronid genomes reveal lophotrochozoan evolution and the origin of bilaterian heads. Nature Ecology & Evolution 2, 141–151 (2018), doi:10.1038/s41559-017-0389-y.


- Inoue, J., Satoh, N. Deuterostome Genomics: Lineage-specific Protein Expansions that Enabled Chordate Muscle Evolution. Molecular Biology and Evolution. 35(4):914-924. (2018)


- Nakano, H., Miyazawa, H., Maeno, A., Shiroishi, T., Kakui, K., Koyanagi, R., Kanda, M., Satoh, N., Omori, A., Kohtsuka, H. Correction to: A new species of Xenoturbella from the western Pacific Ocean and the evolution of Xenoturbella. BMC Evolutionary Biology. 18:83. doi:10.1186/s12862-018-1190-5. (2018)


- Gerdol, M., Luo, Y.J., Satoh, N., Pallavicini, A. Genetic and molecular basis of the immune system in the brachiopod Lingula anatine. Developmental and Comparative Immunology. 82: 7-30. (2018)


- Tominaga, H., Satoh, N., Ueno, N., Takahashi, H. Enhancer activities of amphioxus Brachyury genes in embryos of the ascidian, Ciona intestinalis. genesis. 56(8):e23240. (2018)


- Horie, T., Horie, R., Chen, K., Cao, C., Nakagawa, M., Kusakabe, T.G., Satoh, N., Sasakura, Y., Levine, M. Regulatory cocktail for dopaminergic neurons in a proto-vertebrate identified by whole embryo single cell transcriptomics. Genes and Development, 32(19-20):1297-1302. doi: 10.1101/gad.317669.118. (2018)


- Irie, N., Satoh, N., Kuratani, S. The phylum Vertebrata: a case for zoological recognition Zoological Letters, 2018 4:32. (2018)


- Nakashima, K. A comparative approach to decipher intestinal animal-microbe associations. Microbial Cell. 5: 522-524. (2018)

(b) Environmental Genomics
- Hamada, M., Schröder, K., Bathia, J., Kürn, U., Fraune, S., Khalturina, M., Khalturin, K., Shinzato, C., Satoh, N., Bosch, T.C.G. Metabolic co-dependence drives the evolutionary ancient Hydra-Chlorella symbiosis eLife. 31;7. pii: e35122. doi: https://doi.org/10.1101/234757 (2018)


- Shoguchi, E., Beedessee, G., Tada, I., Hisata, K., Kawashima, T., Takeuchi, T., Arakaki, N., Fujie, M., Koyanagi, R., Roy, M.C., Kawachi, M., Hidaka, M., Satoh, N., Shinzato, C. Two divergent Symbiodinium genomes reveal conservation of a gene cluster for sunscreen biosynthesis and recently lost genes. BMC Genomics. 19:458. doi:10.1186/s12864-018-4857-9. (2018)


- Zayasu, Y., Satoh, N., Shinzato, C. Genetic diversity of farmed and wild populations of the reef building coral, Acropora tenuis. Restoration Ecology. 26(6):1195-1202. doi: 10.1111/rec.12687 (2018)

- Shinzato, C., Zayasu, Y., Kanda, M., Kawamitsu, M., Satoh, N., Yamashita, H., Go Suzuki, G. Using seawater to document coral-zoothanthella diversity: A new approach to coral reef monitoring using environmental DNA
Front. Mar. Sci. 5:28. doi: 10.3389/fmars.2018.00028 (2018)
- Guzman, C., Han, X., Shoguchi, E., Nic Chormaic, S. Fluorescence from a single Symbiodinium cell. Methods Appl Fluoresc. 8(1): 8397. doi: 10.1088/2050-6120/aaba89. (2018)

- Guzman, C., Shinzato, C., Lu, T.M., Conaco, C. Transcriptome analysis of the reef-building octocoral, Heliopora coerulea. Scientific Reports 8:8397. DOI:10.1038/s41598-018-26718-5 (2018)


- Mohamed, A., Cumbo, V., Harii, S., Shinzato, C., Chan, C.X., Ragan, M., Satoh, N., Ball, E., Miller, D. Deciphering the nature of the coral-Chromera association.
The ISME Journal. 12(3): 776-790 (2018)
- Higa, Y., Shinzato, C., Zayasu, Y., Nagata, T., Nakamura, R., Yokokura, A., Shanado, S., Ohmori M. Flexible development of techniques for coral reef restoration using asexual reproduction in the Coral Reef Preservation and Rehabilitation Project by Okinawa Prefectural Government, Japan (in Japanese) Journal of the Japanese Coral Reef Society, 20: 21-37 (2018)

- Mao, Y.F., Economo, E.P., Satoh, N. The roles of introgression and climate change in the rise to dominance of Acropora corals. Current Biology. 28(21):3373-3382.e5. doi: 10.1016/j.cub.2018.08.061. (2018)


- Maeda, K., Saeki, T. Revision of species in Sicyopterus (Gobiidae: Sicydiinae) described by de Beaufort (1912), with a first record of Sicyopterus longifilis from Japan. Species Diversity 23: 253–262 (2018)

- Nakajima, Y., Shinzato, C., Khalturina, M., Nakamura, M., Watanabe, H.K., Nakagawa, S., Satoh, N., Mitarai, S. Isolation and characterization of novel polymorphic microsatellite loci for the deep-sea hydrothermal vent limpet, Lepetodrilus nux, and the vent-associated squat lobster, Shinkaia crosnieri. Marine Biodiversity 48: 677-684. (2018)

- Maeda, K., Saeki, T., Shinzato, C., Koyanagi, R., Satoh, N.
Review of Schismatogobius (Gobiidae) from Japan, with the description of a new species. Ichthyol Res 65: 56-77. (2018)
(c) Functional Genomics
- Shibata H., Chijiwa T., Oda-Ueda N., Nakamura H., Yamaguchi K., Hattori S., Matsubara K., Matsuda Y., Yamashita A., Isomoto A., Mori K., Tashiro K., Kuhara S., Yamasaki S., Fujie M., Goto H., Koyanagi R., Takeuchi T., Fukumaki Y., Ohno M., Shoguchi E., Hisata K., Satoh N., Ogawa T. The habu genome reveals accelerated evolution of venom protein genes. Scientific Reports. 26;8(1):11300. doi: 10.1038/s41598-018-28749-4. (2018)

- Nakashima, K., Kimura, S., Ogawa, Y., Watanabe, S., Soma, S., Kaneko, T., Yamada, L., Sawada, H., Tung, CH., Lu, T.M., Yu, J.K., Villar-Briones, A., Kikuchi, S., Satoh, N.
Chitin-based barrier immunity and its loss predated mucus-colonization by indigenous gut microbiota. Nature Communication. 9(1):3402. doi: 10.1038/s41467-018-05884-0. (2018)

- Matsuura, A., Yoshimura, K., Kintsu, H., Atsumi, T., Tsuchihashi, Y., Takeuchi, T., Satoh, N., Negishi, L., Sakuda, S., Asakura, T., Imura, Y., Yoshimura, E. Structural and functional analyses of calcium ion response factors in the mantle of Pinctada fucata. Journal of Structural Biology. 204(2):240-249. (2018)


- Kawahara, T., Izumikawa, M., Kozone, I., Hashimoto, J., Kagaya, N.,Koiwai, H., Komatsu, M., Fujie, M., Sato, N., Ikeda, H., Shin-ya, K. Neothioviridamide, a Polythioamide Compound Produced by Heterologous Expression of a Streptomyces sp. Cryptic RiPP Biosynthetic Gene Cluster J. Nat. Prod. 81(2):264-269. (2018)

- Iwasaka, H., Koyanagi, R., Satoh, R., Nagano, A., Watanabe, K., Hisata, K., Satoh, N., Aki, T. A Possible Trifunctional β-Carotene Synthase Gene Identified in the Draft Genome of Aurantiochytrium sp. Strain KH105. Genes. 9(4): 200 (2018)


- Tsutsumi, H., Katsuyama, Y., Izumikawa, M., Takagi, M., Fujie, M., Satoh, N., Shin-ya, K., Ohnishi, Y. Unprecedented Cyclization Catalyzed by a Cytochrome P450 in Benzastatin Biosynthesis JACS 140(21):6631-6639. (2018)

- Takeuchi, T., , Plasseraud, L., Ziegler-Devin, I., Brosse, N., Shinzato, C., Satoh, N., Marin, F. Biochemical characterization of the skeletal matrix of the massive coral, Porites australiensis - The saccharide moieties and their localization
Journal of Structural Biology. S1047-8477(18)30135-7. (2018)

- Zhao, R., Takeuchi, T., Luo, Y.J., Ishikawa, A., Kobayashi, T., Koyanagi, R., Villar-Briones, A., Yamada, L., Sawada, H., Iwanaga, S., Nagai, K., Satoh, N., Endo, K.
Dual gene repertoires for larval and adult shells reveal molecules essential for molluscan shell formation. Molecular Biology and Evolution. msy172. https://doi.org/10.1093/molbev/msy172 (2018)

- Kubota, K., Kintsu, H., Matsuura, A., Tsuchihashi, Y., Takeuchi, T., Satoh, N., Suzuki, M. Functional analyses of MMPs for aragonite crystal formation in the ligament of Pinctada fucata. Frontiers in Marine Science, 5:373. doi: 10.3389/fmars.2018.00373 (2018)

- Kim, O.T.P., Nguyen, P.T., Shoguchi, E., Hisata, K., Vo, T.T.B., Inoue, J., Shinzato, C., Le, B.T.N, Nishitsuji, K., Kanda, M., Nguyen, V.H., Nong, H.V., Satoh, N. A draft genome of the striped catfish, Pangasianodon hypophthalmus, for comparative analysis of genes relevant to development and a resource for aquaculture improvement. BMC Genomics, 19(1):733. doi: 10.1186/s12864-018-5079-x. (2018)

- Marin, F., Chmiel, A., Takeuchi, T., Bundeleva, I., Durlet, C., Samankassou, E., Medakovic, D. Skeletal Organic Matrices in Molluscs: Origin, Evolution, Diagenesis. In. Biomineralization. Switzerland: Springer Nature. p. 325-332. (2018)




