FY2022 Annual Report
FY2022 Annual Report
Abstract
The genome contains all the genetic information of a given organism. Decoding the genome provides the molecular basis for understanding every biological phenomenon. Since 2009, the Marine Genomics Unit (MGU) has conducted research in the realm of genome-based biological sciences. By sequencing genomes of target marine organisms (mainly marine invertebrates), we wish to understand genetic and developmental mechanisms underlying evolution and diversification of marine organisms. The major research fields are (a) evolutionary and developmental genomics of marine invertebrates, (b) environmental genomics of coral reefs, and (c) functional genomics of marine organisms including pearl oyster and algae. To date, we have reported draft genomes of a coral in 2011, a pearl oyster in 2012, and coral-symbiotic dinoflagellate (Breviolum) in 2013. We further decoded genomes of hemichordates and a brachiopod in 2015; a brown alga (Okinawa-mozuku) in 2016; Crown-of-Thorns starfish in 2017; a nemertean, phoronid, and two dinoflagellate clades in 2018; jellyfish, dicyemid, acoel flatworm, siphonous macroalga (umi-budo), brown alga (ito-mozuku) in 2019; and hydra and four four strains of “Okinawa mozuku” brown alga in 2020. IN 2021 we have reported sequenced genome of 19 coral species (collaboration with Tokyo Univ.), the kuruma shrinp (collaboration with Tokyo U. of Marine Sci. Tech.), and nearly complete genome of the tunicate Ciona (collaboration with Kyoto U.). In this year, we reported haplotype-phased genome of a pearl oyster . In addition, we have advanced genome-based coral research, especialy coral-specific eDNA projects, and one research result shall be reported below.

[Photo on October 12, 2022]
1. Staffs and Students
- Professor Noriyuki Satoh
- Staff Scientists
- Eiichi Shoguchi (Group Leader)
- Keisuke Nakashima
- Konstantin Khalturin (~Sept. 2022)
- Takeshi Takeuchi
- Koki Nishitsuji
- Takeshi Noda
- Technical Staffs
- Kanako Hisata
- Sakura Kikuchi
- Haruhi Narisoko
- Research Assistants
- Yoshie Nishitsuji (part time)
- Mayuki Suwa (part time)
- Research Administrators
- Shoko Yamakawa
- Tomomi Teruya
- Students
- Ph.D students (co-superviser)
- Rio Kashimoto (Supervisor: Prof. Laudet, V.)
- Ph.D students (co-superviser)
2. Collaborations
2-1 Genome scientific studies of chordate evolution
- Type of collaboration: Scientific collaboration
- Researchers: Prof. Daniel Rokshar, OIST
2-2 Genome sequencing of marine invertebrates at haplotype-resolution level
- Type of collaboration: Scientific collaboration
- Researchers: Prof. Gene Myers, OIST
2-3. Molecular biological study of COTS communications
- Type of collaboration: Scientific collaboration
- Researchers: Prof. Scott Cummins, Univ. Sunshine Coast, Australia
2-4. Genome scientific study of dinoflagellates
- Type of collaboration: Scientific collaboration
- Researchers: Prof. Pengchen Fu, Hainan University
2-5. Genome scientific study of coral-dinoflagellate symbiosis
- Type of collaboration: Scientific collaboration
- Researchers: Profs. Shigeki Fujiwara & Kaz Kawamura, Kochi University
2-6. Genome scientific study of left-right asymmetry of snails
- Type of collaboration: Scientific collaboration
- Researchers: Prof. Takehiro Asami, Shinshu University
2-7. Genome scientific study of acoel development
- Type of collaboration: Scientific collaboration
- Researchers: Profs. Kunifumi Tagawa, Asuka Arimoto & Tatsuya Ueki, Hiroshima University
3. Research activities and findings
An environmental DNA metabarcoding survey reveals generic-level occurrence of scleractinian corals at reef slopes of Okinawa Island
Proc. Roy. Soc. B, 290: 20230026
Coral reefs have the highest biodiversity of all marine ecosystems in tropical and subtropical oceans. However, scleractinian corals, keystone organisms of reef productivity, are facing a crisis due to climate change and anthropogenic activities. A broad survey of reef-building corals is essential for world-wide reef preservation. To this end, direct observations made by coral-specialist divers might be supported by another robust method. We improved a recently devised environmental DNA (eDNA) metabarcoding method by out team to identify more than 43 scleractinian genera by sampling 2 L of surface seawater above reefs (Shinzato et al., 2021, Front. Marine Sci. 8, 758207).
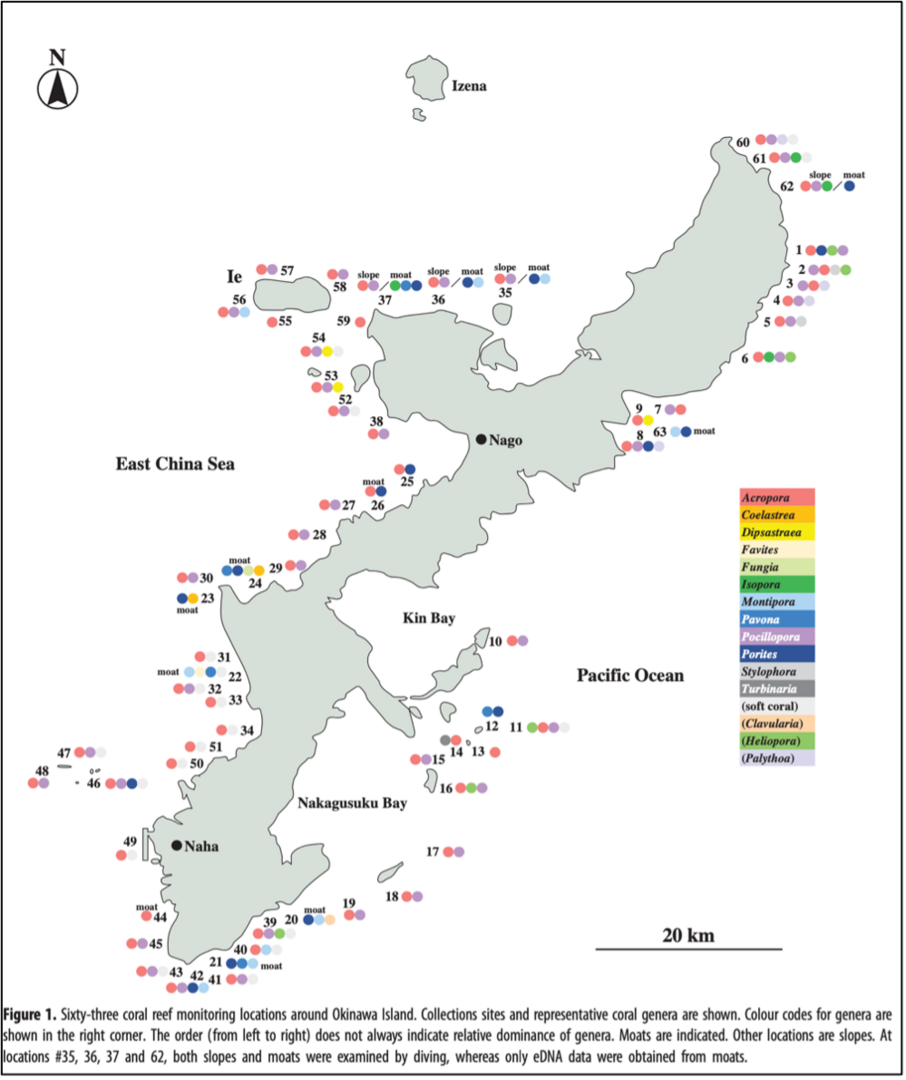
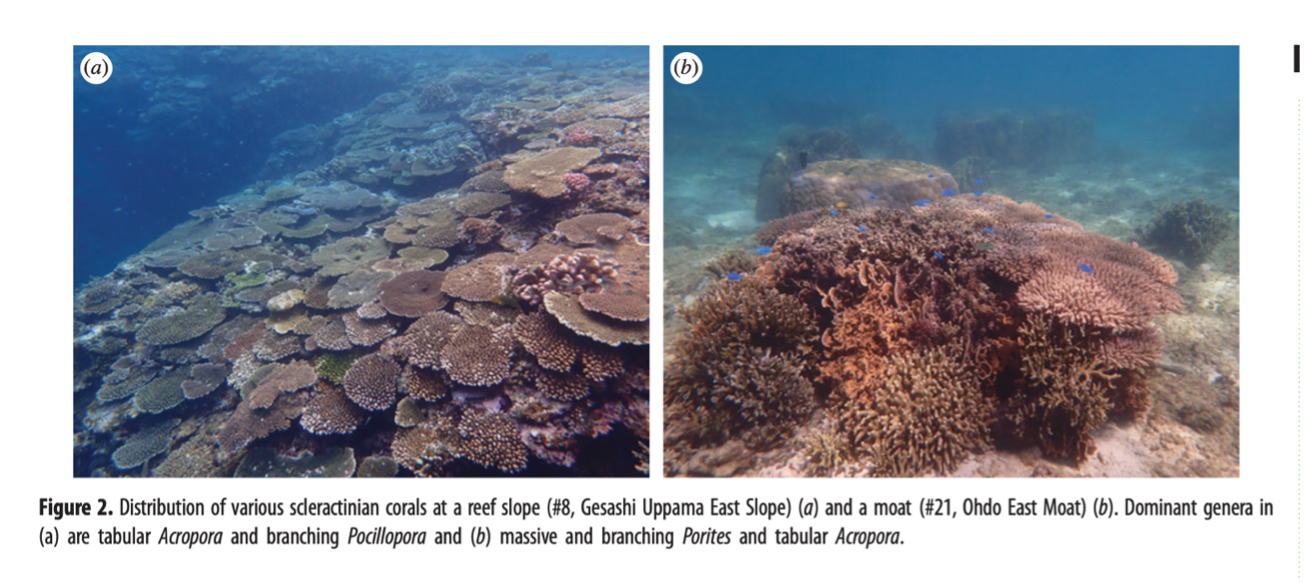
In this study, together with direct observations by divers (a part of Monitoring Sites 1000 Project supported by the Ministry of the Environment of Japan), we assessed the utility of eDNA at 63 locations spanning ~250 km near Okinawa Island (Fig. 1). Of 63 monitoring sites, 51 were slopes (3-10m in depth; Fig. 2a), 8 were moats (1-3m in depth; Fig. 2b), and 4 sites were both slopes and moats (#35-37, 62).
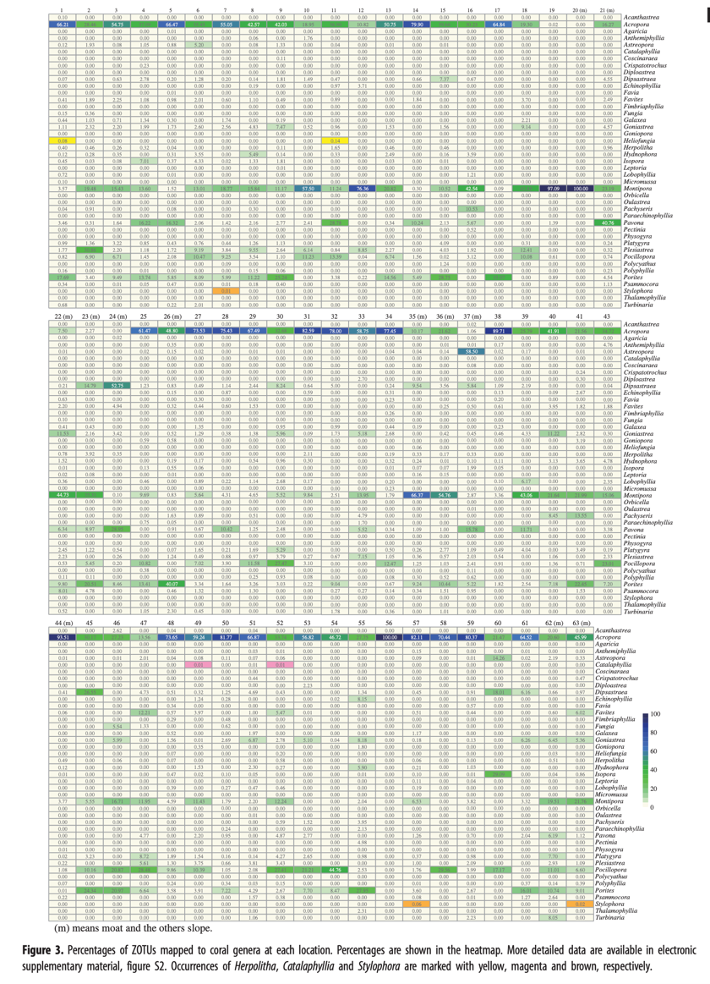
Major genera recorded by divers included Acropora, Pocillopora, Porites, and Montipora (Fig. 1). Simultaneously, scleractinian coral-specific eDNA barcoding analyses were carried out at 62 locations. We obtained amplicons from all samples at 62 sites, confirming that 2 L of surface seawater are enough for scleractinian-specific eDNA metabarcoding (Fig. 3). Results of the scleractinian coral-specific eDNA barcoding analyses confirmed genera recorded by direct observations by divers.
In addition, the eDNA method identified more genera than direct observations and documented the presence of previously unrecorded species. For example, eDNA likely provided evidence for genera that were so far unknown from Okinawa Island. For example, two species in the genus Heliofungia have been reported , H. actiniformis (Quoy and Gaimard 1833) and H. fralinae (Nemenzo 1959), the latter being recorded in southern Asia, including Japan. However, this species reportedly reaches its northern limit at Miyako Island, and there have been no reports of its presence at Okinawa or Amami Islands (Fig. 3). Therefore, it was suggest a more northward distribution of H. fralinae, reaching at least as far as Okinawa Island.
Finally, to evaluate this scleractinian coral-specific eDNA method, we compared results of eDNA barcoding with direct observations (Table 1). As a result, 41 of 62 points were well matched (67%), 15 were moderately matched (24%), 4 were partially match (6%), and only 2 points showed no match (3%). In other words, at more than 91% (67%+24%) of monitored locations, eDNA results were confirmed by direct observations.
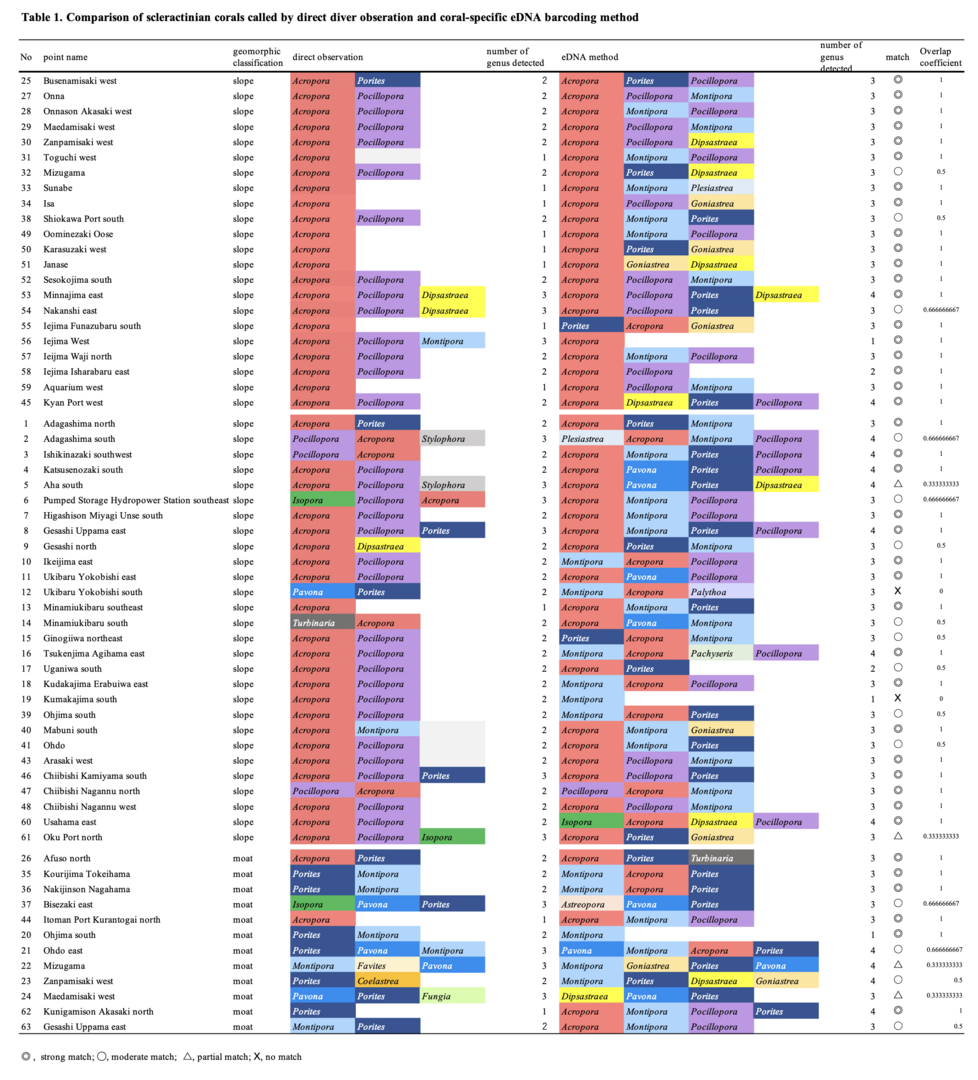
Therefore, we concluded that this scleractinian coral-specific eDNA method promises to be a powerful tool to survey coral reefs broadly, deeply, and robustly.
4. Publications
(a) Developmental and Evolutionary Genomics
- Khalturin, K., Shunatova, N., Shchenkov, S., Sasakura, Y., Kawamitsu, M., Satoh, N.
Polyzoa is back: The effect of complete gene sets on the placement of Ectoprocta and Entoprocta
Science Advances, 8(26):eabo4400. (2022)
- Satoh, N., Hisata, K., Foster, S., Morita, S., Nishitsuji, K., Oulhen, N., Tominaga, H., Wessel, G.
A single-cell RNA-seq analysis of Brachyury-expressing cell clusters suggests a morphogenesis-associated signal center of oral ectoderm in sea urchin embryos
Developmental Biology, 483: 128-142 (2022)
- Humphreys, T., Weiser, K., Arimoto, A., Sasaki, A., Uenishi, G., Fujimoto, B., Kawashima, T., Taparra, K., Molnar, J., Satoh, N., Marikawa, Y., Tagawa, K.
Ancestral stem cell reprogramming genes active in hemichordate regeneration
Frontiers in Ecology and Evolution, 10: 769433 (2022)
- Kashimoto, R., Tanimoto, M., Miura, S., Satoh, N., Laudet, V., Khalturin, K.
Transcriptomes of giant sea anemones from Okinawa as a tool for understanding their phylogeny and symbiotic relationships with anemonefish
Zoological Science, 39(4) (2022)

(b) Environmental Genomics
- Nishitsuji, K., Nagata, T., Narisoko, H., Kanai, M., Hisata, K., Shinzato, C., Satoh, N.
An environmental DNA metabarcoding survey reveals generic-level occurrence of scleractinian corals at reef slopes of Okinawa Island.
Proceedings B, 290:20230026 (2023)
- Yasuda, N., Inoue, J., Hall, M.R., Nair, M.R., Adjeroud, M., Miguel D. Fortes, M.D., Nishida, M., Tuivavalagi, N., Ravago-Gotanco, R., Forsman, Z.H., Soliman, T., Koyanagi, R., Hisata, K., Motti, C.A., Satoh, N.
Two Hidden mtDNA-Clades of Crown-of-Thorns Starfish in the Pacific Ocean
Frontiers Marine Science, 9: 831240, doi: 10.3389/fmars.2022.831240 (2022)
- Shimakawa, G., Shoguchi, E., Burlacot, A., Ifuku, K., Che, Y., Kumazawa, M., Tanaka, K., Nakanishi, S.
Coral symbionts evolved a functional polycistronic flavodiiron gene.
Photosynth Res. 151(1):113-124. doi: 10.1007/s11120-021-00867-7. (2022)
- Tsuchiya, K., Zayasu, Y., Nakajima, Y., Arakaki, N., Suzuki, G., Satoh, N., Shinzato, C.
Genomic analysis of a reef-building coral, Acropora digitifera, reveals complex population structure and a migration network in the Nansei Islands, Japan
Molecular Ecology, 31, 5270– 5284. (2022)
- Kitchen, S.,A., Jiang, D., Harii, S., Satoh, N., Weis, V.M., and Shinzato, C.
Coral larvae suppress the heat stress response during the onset of symbiosis thereby decreasing their odds of survival
Molecular Ecology, 31:5813-5830. (2022)
- Kiyono, S., Miyashita, S., Yamaguchi, K., Saito, I., Saito, Y., Manta, S., Ishikawa, M., Narita, M., Watanabe, T., Ito, R., Taguchi, M., Furukawa, R., Ikeuchi, A., Matsuo, K., Kurita, G., Kumagai, T., Shirakashi, S., Ogawa, K., Sakamoto, K., Koyanagi, R., Satoh, N., Sasaki, M., Maezawa, T., Ichikawa-Seki, M., Kobayashi. K.
Sex-inducing effects toward planarians widely present among parasitic flatworms
iScience, 26:105776 (2022)

(c) Functional Genomics
- Takeuchi, T., Suzuki, Y., Watabe, S., Nagai, K., Tetsuji Masaoka, T., Fujie, M., Kawamitsu, M., Satoh, N., Myers, E.W.
A high-quality, haplotype-phased genome reconstruction reveals unexpected 3 haplotype diversity in a pearl oyster
DNA Research, 29:dsac035. (2022)
- Isomoto, A., Shoguchi, E., Hisata, K., Inoue, J., Inaba, K., Satoh, N., Ogawa, T., Hiroki Shibata, H.
Active Expression of Genes for Protein Modification Enzymes in Habu Venom Glands
toxins, 14, 300. (2022)

- Nishitsuji, K., Nishitsuji, Y., Yonashiro, Y., Satoh, N.
Development of DNA markers that distinguish male and female haploid germlings of the brown alga, Cladosiphon okamuranus
Phycological Research, 70: 160-166.
- Shimizu, K., Takeuchi, T., Negishi, L., Kurumizaka, H., Kuriyama, I., Endo, K., Suzuki, M.
Evolution of Epidermal Growth Factor (EGF)-like and Zona Pellucida Domains Containing Shell Matrix Proteins in Mollusks.
Mol Biol Evol. 39(7):msac148. doi: 10.1093/molbev/msac148. (2022)

- Adi, T.K., Fujie, M., Satoh, N., Ueki, T.
The acidic amino acid-rich C-terminal domain of VanabinX enhances reductase activity, attaining 1.3- to 1.7-fold vanadium reduction
Biochemistry and Biophysics Reports, 32, 101349 (2022)

- Boutet, I., Alves Monteiro, H. J., Baudry, L., Takeuchi, T., Bonnivard, E., Billoud, B., Farhat, S., Gonzales-Araya, R., Salaun, B., Andersen, A. C., Toullec, J.-Y., Lallier, F. H., Flot, J.-F., Guiglielmoni, N., Guo, X., Li, C., Allam, B., Pales-Espinosa, E., Hemmer-Hansen, J., Moreau, P., Marbouty, M., Koszul, R., Tanguy, A.
Chromosomal assembly of the flat oyster (Ostrea edulis L.) genome as a new genetic resource for aquaculture
Evolutionary Applications, 00, 1– 19.
(d) Sequencing Collaboration
- Yamamoto, K., Yoneda, Y., Makino, A., Tanaka, Y., Meng, X.Y, Hashimoto, Y., Shin-ya, K., Satoh, N., Fujie, M., Toyama, T., Mori, K., Ike, M., Morikawa, M., Kamagata, Y., and Tamaki H.
Draft genome sequence of Bryobacteraceae strain F-183
Microbiology Resource Announcements, 11(1):e0045321. doi: 10.1128/mra.00453-21. (2022)
- Yamamoto, K., Yoneda, Y., Makino, A., Tanaka, Y., Meng, X.Y, Hashimoto, Y., Shin-ya, K., Satoh, N., Fujie, M., Toyama, T., Mori, K., Ike, M., Morikawa, M., Kamagata, Y., and Tamaki H.
Complete Genome Sequence of Luteitalea sp. Strain TBR-22.
Microbiol Resour Announc. 11(2):e0045521. doi: 10.1128/mra.00455-21. (2022)
- Choirunnisa, A. R., Arima, K., Abe, Y., Kagaya, N., Kudo, K., Suenaga, H., Hashimoto, J., Fujie, M., Satoh, N., Shin-ya, K., Matsuda, K., Wakimoto, T.
New azodyrecins identified by a genome mining-directed reactivity-based screening
Beilstein J. Org. Chem. 18, 1017–1025. https://doi.org/10.3762/bjoc.18.102

5. Seminar
- Dr. Yuuri Yasuoka
Researcher in RIKEN Center for Integrative Medical Sciences
Title: "Tissue-specific expression of carbohydrate sulfotransferases drives keratan sulfate biosynthesis in the notochord and otic vesicles of Xenopus embryos"
Date: October 5th, 2022 - 13:00 to 14:00
Venue: D015 - Dr. Christopher B. Cameron
Professor in the Department of Biological Sciences at University of Montreal.
Title: "The origin of gills, skeletal ossicles & tubes in the deuterostomes"
Date: November 21st, 2022 – 16:00 to 17:00
Venue: C210



