FY2019 Annual Report

[July 25, 2019]
Abstract
The genome contains all the genetic information of a given organism. Decoding the genome therefore provides the molecular basis for understanding every biological phenomenon. Since 2009, the Marine Genomics Unit (MGU) has conducted research in the realm of genome-based biological sciences. By decoding genomes of target marine organisms (mainly invertebrates), we wish to understand genetic and developmental mechanisms underlying diversity of marine organisms. The major research fields are (a) evolutionary and developmental genomics of marine invertebrates, (b) environmental genomics of coral reefs, and (c) functional genomics of marine organisms, including pearl oyster and algae. To date, we have decoded genome of a coral in 2011, a pearl oyster in 2012, and symbiotic dinoflagellate (Symbiodinium) in 2013. We further decoded genomes of hemichordates and a brachiopod in 2015, a brown alga (Okinawa-mozuku) in 2016, Crown-Thorns-Starfish in 2017, and a nemertean, phoronid, and two dinoflagellate clades in 2018. This year we deciphered genomes of two jellyfish, dicyemid, acoel flatworm, siphonous macroalga (umi-budo), and brown alga (ito-mozuku). We also improved the Ciona intestinalis genome to be nearly complete one. Main results of this year research shall be reported below.
1. Staff FY2019
- Professor Noriyuki Satoh
- Staff Scientists
- Eiichi Shoguchi (Group Leader)
- Keisuke Nakashima
- Konstantin Khalturin
- Ken Maeda
- Takeshi Takeuchi
- Koki Nishitsuji
- Yuna Zayasu
- Postdoctoral Scholars
- Asuka Arimoto (~6/30, '19)
- Hitoshi Tominaga
- JSPS Fellow
- Remi N. Ketchum (University of North Carolina at Charlotte)
- PhD Students
- (Superviser)
- Girish Beedessee (JSPS DC Fellow)
- Yafei Mao (JSPS DC Fellow)
- (co-superviser)
- Kun-Lung Li (Supervisor: Assistant Prof. Watanabe, H.)
- Christina Ripken (Supervisor: Prof. Shannon, N.)
- Lab rotation students
- Billy Moore
- Rio Kashimoto
- Internship Students
- Yang Shu (Hainan Univ, China)
- Aya Koseki (Kitasato Univ)
- (Superviser)
- Technical Staffs
- Kanako Hisata
- Sakura Kikuchi
- Haruhi Narisoko
- Research Assistants
- Seiya Kitanobo (POC Program)
- Yoshie Nishitsuji (AIMS project)
- Mayuki Suwa
- Wakana Chinen (Sep,2019 ~ Mar, 2020)
- Research Administrators
- Shoko Yamakawa
- Tomomi Teruya
2. OIST PhD Graduation
We congratulate a student for his OIST PhD Graduation.
3. The Research Encouragement Prize of Okinawa 2019

Dr. Koki Nishitsuji(middle left) and Dr. Asuka Arimoto(left) were awarded the Research Encouragement Prize of Okinawa 2019 as recognition for their contribution to Okinawa for their work on deciphering the genomes of many algal crops, including Okinawa mozuku and Umi-budo.
4. Collaborations
4-1 Genome scientific studies of chordate evolution
- Type of collaboration: Scientific collaboration
- Researchers: Prof. Daniel Rokshar, OIST
4-2. Molecular biological study of COTS communications
- Type of collaboration: Scientific collaboration
- Researchers: Prof. Scott Cummins, Univ. Sunshine Coast, Australia
4-3. Genome scientific study of dinoflagellates
- Type of collaboration: Scientific collaboration
- Researchers: Prof. Pengchen Fu, Hainan University
4-4. Genome scientific study of coral-dinoflagellate symbiosis
- Type of collaboration: Scientific collaboration
- Researchers: Profs. Shigeki Fujiwara & Kaz Kawamura, Kochi University
4-5. Genome scientific study of left-right asymmetry of snails
- Type of collaboration: Scientific collaboration
- Researchers: Prof. Takehiro Asami, Shinshu University
4-6. Genome scientific study of acoel development
- Type of collaboration: Scientific collaboration
- Researchers: Profs. Kunifumi Tagawa & Tatsuya Ueki, Hiroshima University
5. Research activities and findings
5.1 Developmental and evolutionary genomics
This year our Unit has deciphered genomes of two jellyfish, dicyemid, acoel flatworm, siphonous macroalga (umi-budo), and brown alga (ito-mozuku). We have also improved the Ciona intestinalis genome to be nearly complete one. Here, we report results of jellyfish genomes, which was published in Nature Ecology and Evolution 3:811-822 (2019) with cover photo of the journal.
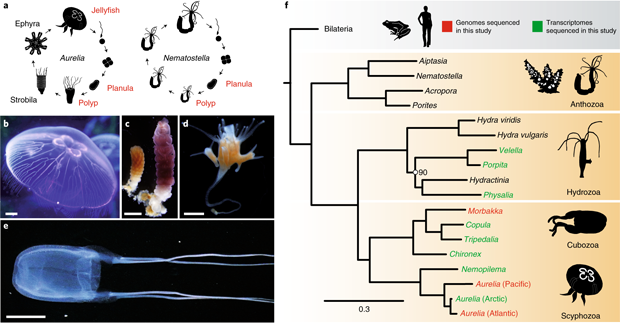
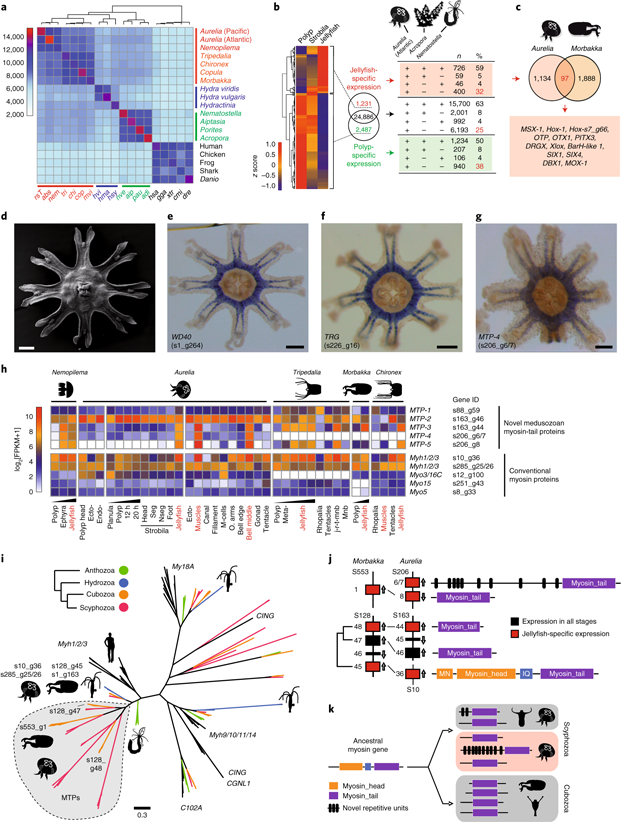
The Cnidaria is an ancient phylum considered a sister group to all bilaterian animals. They are astonishingly diverse in body form and lifestyle, including the presence of a jellyfish stage in medusozoans and its absence in anthozoans (Fig. 1). Here, we sequence the genomes of Aurelia aurita (a scyphozoan) and Morbakka virulenta (a cubozoan) to understand the molecular mechanisms responsible for the origin of the jellyfish body plan. We show that the magnitude of genetic differences between the two jellyfish types is equivalent, on average, to the level of genetic differences between humans and sea urchins in the bilaterian lineage. About one-third of Aurelia genes with jellyfish-specific expression have no matches in the genomes of the coral and sea anemone, indicating that the polyp-to-jellyfish transition requires a combination of conserved and novel, medusozoa-specific genes (Fig. 2). While no genomic region is specifically associated with the ability to produce a jellyfish stage, the arrangement of genes involved in the development of a nematocyte—a phylum-specific cell type—is highly structured and conserved in cnidarian genomes; thus, it represents a phylotypic gene cluster.
5.2 Functional genomics
We report here results of decoded genome of the siphonous green alga, Caulerpa lentillifera, which was published in DNA Research 26:183-192 (2019). Genome evolution and development of unicellular, multinucleate macroalgae (siphonous algae) are poorly known, although various multicellular organisms have been studied extensively. To understand macroalgal developmental evolution, we assembled approximately 26Mb genome of a siphonous green alga, Caulerpa lentillifera (Fig. 1), with high contiguity, containing 9,311 protein- coding genes. Molecular phylogeny using 107 nuclear genes indicates that the diversification of the class Ulvophyceae, including C. lentillifera, occurred before the split of the Chlorophyceae and Trebouxiophyceae. Compared with other green algae, the TALE superclass of homeobox genes, which expanded in land plants, shows a series of lineage-specific duplications in this siphonous macroalga (Fig. 2). Plant hormone signalling components were also expanded in a lineage- specific manner. Expanded transport regulators, which show spatially different expression, suggest that the structural patterning strategy of a multinucleate cell depends on diversification of nuclear pore proteins. These results not only imply functional convergence of duplicated genes among green plants, but also provide insight into evolutionary roots of green plants. Based on the present results, we propose cellular and molecular mechanisms involved in the structural differentiation in the siphonous alga.
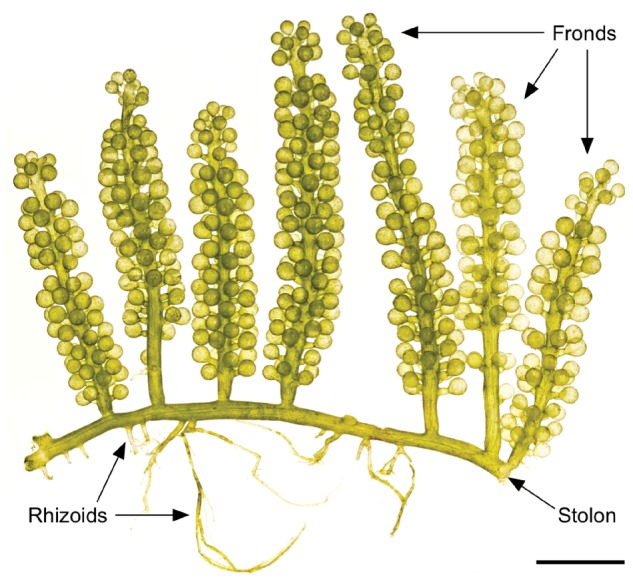
Figure 1 | The siphonous alga, Caulerpa lentillifera. Cultivated C. lentillifera (photo by Dr Ken Maeda). The alga consists of many grape-like vesicles connected by stolons, and the entire alga comprises one cell with many nuclei. Scale bar, 10 mm.
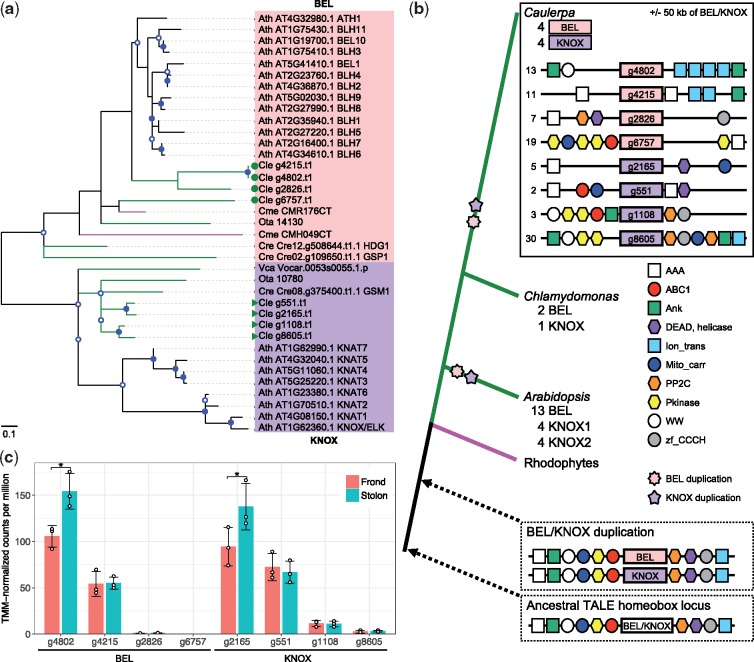
Figure 2 | Molecular phylogenetic analysis showing lineage-specific evolution of TALE class homeobox proteins in green plants and the ancient plant genomic structure. (a) The maximum-likelihood tree is constructed using multiple alignments of homeodomains. Green symbols show four members of the KNOX class and four of the BEL class in the Caulerpa genome. Nodes with more than 50 and 70% bootstrap support are marked with open and closed circles, respectively. Green, magenta and black lines correspond to chlorophyte, rhodophyte and Arabidopsis sequences, respectively. Ath, Arabidopsis thaliana. Cle, Caulerpa lentillifera. Cme, Cyanidioschyzon merolae. Cre, Chlamydomonas reinhardtii. Ota, Ostreococcus tauri. Vca, Volvox carteri. (b) Origin and evolution of the plant BEL/KNOX family and candidate genes for exploring the duplication mechanism of the ancestral gene. Simplified gene order in the Caulerpa genome is shown above. Scaffold numbers are indicated at the left side. Potential ancestral genomic structures are surrounded by dotted lines. Circles, rectangles and hexagonal boxes in diagrams of genomic sequences indicate protein domains contained in each locus. The synteny diagrams are not drawn to scale. (c) Expression levels of TALE homeobox genes. * indicates P-value <0.05 and false discovery rate <0.05. Two genes, g4802 of BEL, and g2165 of KNOX, show the difference of gene expression level between frond and stolon. Error bars and white circles show standard deviation and TMM-normalized CPM observed in each replicate, respectively.
6. Publications
(a) Developmental and Evolutionary Genomics
- Satoh, N.
A deep dive into sea-squirt development
Nature 571, 333-334 (2019)

- Arimoto, A., Hikosaka-Katayama, T., Hikosaka, A., Tagawa, K., Inoue, T., Ueki, T., Yoshida, M., Kanda, M., Shoguchi, E., Hisata, K., Satoh, N.
A draft genome assembly of the acoel flatworm Praesagittifera naikaiensis.
GigaScience, 8(4) (2019)
- Lu, T.M., Kanda, M., Furuya, H., Satoh, N.
Dicyemid mesozoans: a unique parasitic lifestyle and a reduced genome
Genome Biology and Evolution, 11:2232–2243 (2019)
- Inoue, J., Satoh, N.
ORTHOSCOPE: an automatic web tool for phylogenetically inferring bilaterian orthogroups with user-selected taxa
Molecular Biology and Evolution, 36: 621-631 (2019)
- Inoue, J., Nakashima, K., Satoh, N.
ORTHOSCOPE Analysis Reveals the Presence of the Cellulose Synthase Gene in All Tunicate Genomes but not in Other Animal Genomes
Genes, 10: 294 (2019)

- Noda, T., Satoh, N., Asami, T.
Heterochirality results from reduction of maternal diaph expression in a terrestrial pulmonate snail
Zoological Letters, 2019 5:2 (2019).

- Marlétaz, F., Peijnenburg, K.T.C.A., Goto, T., Satoh, N., Rokhsar, D.S.
A new spiralian phylogeny places the enigmatic arrow worms among gnathiferans
Current Biology, 29, 1–7. (2019)

- Philippe, H., Poustka, A.J., Chiodin, M., Hoff, K.J., Dessimoz, C., Tomiczek, B., Schiffer, P.H., Muller, S., Domman, D., Horn, M., Kuhl, H., Timmermann, B., Satoh, N., Hikosaka-Katayama, T., Nakano, H., Rowe, M.L., Elphick, M.R., Thomas-Chollier, M., Hankeln, T., Mertes, F., Wallberg, A., Rast, J.P., Copley, R.R., Martinez, P., Telford M.J.
Mitigating Anticipated Effects of Systematic Errors Supports Sister-Group Relationship between Xenacoelomorpha and Ambulacraria
Current Biology, 29: 1818-1826 (2019)

- Ueki, T., Arimoto, A., Tagawa, K., Satoh, N.
Xenacoelomorph-Specific Hox Peptides: Insights into the Phylogeny of Acoels, Nemertodermatids, and Xenoturbellids
Zoological Science, 36: 395-401. (2019)
- Ishikawa, A., Kabeya, N., Ikeya, K., Kakioka, R., Cech, J.N., Osada, N., Leal, M.C., Inoue, J., Kume, M., Toyoda, A., Tezuka, A., Nagano, A.J., Yamasaki, Y.Y,. Suzuki, Y., Kokita, T., Takahashi, H., Lucek, K., Marques, D., Takehana, Y., Naruse, K., Mori, S., Monroig, O., Ladd, N., Schubert, C., Matthews, B., Peichel, C.L., Seehausen, O., Yoshizaki, G., Kitano, J.
A key metabolic gene for recurrent freshwater colonization and radiation in fishes
Science, 364: 886-889 (2019)

- Shiraishi, A., Okuda, T., Miyasaka, N., Osugi, T., Okuno, Y., Inoue, J., & Satake, H.
Repertoires of G protein-coupled receptors for Ciona-specific neuropeptides.
PNAS, 116: 7847–7856. doi:10.1073/pnas.1816640116 (2019)

- Yasuoka, Y., Tando, Y., Kubokawa, K., Taira, M.
Evolution of cis-regulatory modules for the head organizer gene goosecoid in chordates: comparisons between Branchiostoma and Xenopus
Zoological Letters, 5: 27 (2019)

- Lu, T.M., Furuya, H., Satoh, N.
Gene expression profiles of dicyemid life-cycle stages may explain how dispersing larvae locate new hosts.
Zoological Letters, 5: 32 (2019)
- Maiorova, M.A., Satoh, N., Khalturin, K., Odintsova, N.A.
Transcriptomic profiling of the mussel Mytilus trossulus with a special emphasis on integrin-like genes during development
Invertebrate Reproduction & Development, 63: 231-240 (2019)
- Satou, Y., Nakmura, R., Deli, Y., Yoshida, R., Hamada, M., Fujie, M., Hisata, K., Takeda, H., Satoh, N.
A nearly-complete genome of Ciona intestinalis type A (C. robusta) reveals the contribution of inversion to chromosomal evolution in the genus Ciona.
Genome Biology and Evolution, 1;11:3144-3157 (2020)

(b) Enviromental Genomics
- Zayasu, Y., Suzuki, G.
Comparisons of population density and genetic diversity in artificial and wild populations of an arborescent coral, Acropora yongei: implications for the efficacy of "artificial spawning hotspots"
Restoration Ecology, https://doi.org/10.1111/rec.12857
- Mao, Y., Satoh, N.
A likely ancient genome duplication in the speciose reef-building coral genus, Acropora
iScience, 13: 20-32 (2019)

- Miyake, T., Aihara, N., Maeda, K., Shinzato, C., Koyanagi, R., Kobayashi, H., Yamahira, K.
Bloodmeal host identification with inferences to feeding habits of a fish-fed mosquito, Aedes baisasi
Scientific Reports, 9: 4002 (2019)

- Hanahara, N., Higashiji, T., Shinzato, C., Koyanagi, R., Maeda, K.
First record of Larsonella pumilus (Teleostei: Gobiidae) from Japan, with phylogenetic placement of the genus Larsonella
Zootaxa 4695: 367-377 (2019)
(c) Functional Genomics
- Beedessee, G., Hisata, K., Roy, M.C., Van Dolah, F.M., Satoh, N., Shoguchi, E.
Diversified secondary metabolite biosynthesis gene repertoire revealed in symbiotic dinoflagellates
Scientific Reports, 9: 1204 (2019)

- Ueki, T., Fujie, M., Romaidi, Satoh, N.
Symbiotic bacteria associated with Ascidian vanadium accumulation identified by 16S rRNA amplicon Sequencing
Marine Genomics, 43: 33-42 (2019)

- Arimoto, A., Nishitsuji, K., Arakaki, N., Hisata, K., Shinzato, C., Satoh, N., Shoguchi, E.
A siphonous macroalgal genome suggests convergent functions of homeobox genes in algae and land plants.
DNA Research, 26: 183–192 (2019)
- Nishitsuji, K., Arimoto, A., Higa, Y., Mekaru, M., Kawamitsu, M., Satoh, N., Shoguchi, E.
Draft genome of the brown alga, Nemacystus decipiens, Onna-1 strain: Fusion of genes involved in the sulfated fucan biosynthesis pathway
Scientific Reports, 9: 4607 (2019)
- Arimoto, A., Nishitsuji, K., Narisoko, H., Shoguchi, E., Satoh, N.
Differential gene expression in fronds and stolons of the siphonous macroalga, Caulerpa lentillifera
Dev. Growth Diff, doi:10.1111/dgd.12634 (2019)

- Ogawa, T., Oda-Ueda, N., Hisata, K., Nakamura, H., Chijiwa, T., Hattori, S., Isomoto, A., Yugeta, H., Yamasaki, S., Fukumaki, Y., Ohno, M., Satoh, N., Shibata, S.
Extensive alternative mRNA splicing underlying highly divergent venom proteins of the habu snake Protobothrops flavoviridis
Toxins, 11,581 (2019)

(d) Other Fields
- Nakashima, K., Kikuchi, S.
Chitin-based barrier immunity and its loss predated mucus-colonization by indigenous gut microbiota. (in Japanese)
実験医学37, 69-72 (2019) - Arimoto, A.
Exploring the mysteries of morphogenesis of sea grapes: insights from the sea grape genome. (in Japanese)
academist journal (2019)
7. Seminar
Dr. Henry Gee
Nature Senior Editor
Title: "What Happens Inside 'Nature'"
Date: January 6th, 2020 - 15:00 to 16:00




 Mao Yafei
Mao Yafei Girish Beedessee
Girish Beedessee
