FY2014 Annual Report
Computational Neuroscience Unit
Professor Erik De Schutter

Abstract
We use computational, data-driven methods to study how neurons and microcircuits in the brain operate. We are interested in how fundamental properties, such as a neuron’s morphology and its excitability, interact with one another during common neural functions like information processing or learning. Most of our models concern the cerebellum as this brain structure has a relatively simple anatomy and the physiology of its main neurons has been studied extensively, allowing for detailed modeling at many different levels of complexity.
1. Staff
General services and neuroinformatics
- Ivan Raikov, Technical Staff (till October 2014)
- Sachie Matsuoka, Research Administrator/ Secretary
Molecular modeling
- Weiliang Chen, Researcher
- Himanshu Gangal, Technical Staff (from June 2014)
- Iain Hepburn, Technical Staff
- Nikon Rasumov, Researcher
Cellular modeling
- Sungho Hong, Group Leader
- Akira Takashima, Researcher
- Yunliang Zang, Researcher (from May 2014)
Network modeling
- Benjamin Torben-Nielsen, Group Leader
- Tom Close, Researcher
- Shyam Kumar Sudhakar, Special Research Student
JSPS Research Fellow
-
Richard Tomsett, Researcher
Research Interns
- Russell Jarvis, Research Intern (from January 2015)
- Chihiro Kurosawa, Research Intern (from February 2015)
2. Collaborations
- Theme: Cerebellar physiology, multiple themes
- Type of collaboration: Scientific collaboration and graduate program
- Researchers:
- Professor M. Giugliano, University of Antwerp, Belgium
- Professor D. Snyders, University of Antwerp, Belgium
- Joao Couto, University of Antwerp, Belgium
- Q. Robberecht, University of Antwerp, Belgium
- Theme: Spiking activity of monkey cerebellar neurons
- Type of collaboration: Scientific collaboration
- Researchers:
- Professor H.P. Thier, University of Tübingen, Germany
- A. Ignashchenkova, University of Tübingen, Germany
- Dr. M. Junker, University of Tübingen, Germany
- A. Schmigdlin, University of Tübingen, Germany
- Theme: Human Brain Project: simulator development
- Type of collaboration: Scientific collaboration
- Researchers:
- Prof. F. Schürmann, École Polytechnique Fédérale de Lausanne, Switzerland
- F. Delalondre, École Polytechnique Fédérale de Lausanne, Switzerland
- Theme: Molecular identification of cerebellar signaling pathways and cerebellar optogenetics
- Type of collaboration: Scientific collaboration
- Researchers:
- Professor K. Tanaka, Korea Institute for Science and Technology (KIST), Korea
- Professor K. Tanaka, Korea Institute for Science and Technology (KIST), Korea
- Theme: Modeling of effects of ethanol on the cerebellum
- Type of collaboration: Joint research
- Researchers:
- Professor C.F. Valenzuela, University of New Mexico, United States of America
- Professor C.F. Valenzuela, University of New Mexico, United States of America
- Theme: Correlation of neurons linked to their excitability
- Type of collaboration: Scientific collaboration
- Researchers:
- Professor S.A. Prescott, University of Toronto, Canada
- Dr. S. Ratté, University of Toronto, Canada
- Theme: Purkinje cell morphology and physiology, modeling
- Type of collaboration: Scientific collaboration
- Researchers:
- Professor M. Häusser, University College London, United Kingdom
- Professor H. Cuntz, Goethe University Germany
- Professor A. Watt, McGill University, Canada
- Dr. A. Roth, University College London, United Kingdom
3. Activities and Findings
3.1 Cellular mechanisms regulating firing and synaptic properties of neurons
Analysis of neural morphologies
We designed and implemented BTMORPH, a Python library to efficiently represent and analyze neuronal morphologies (Torben-Nielsen, 2014). The rationale of this library is to provide a solid, well tested backbone in the form of a data structure and atomic morphometric functions that allow users to analyze morphologies in a flexible way. Neuronal morphologies are treated as tree structures and all provided morphometrics can be computed on any (sub)tree structure. As such, morphometrics can be performed on the whole structure (as is usually done) or on any subtree; subtrees can be trees made up by a specific neurite type (axon, apical dendrite, ...) or can be selected specifically by, for instance, centrifugal order.
The target audience of BTMORPH consists of researchers who can read and modify Python scripts. However, we also provide straightforward wrappers around the code to perform essential functionality such as analyzing multi- dimensional (e.g., conditional) data in single neurons or to record population statistics. A typical example of using BTMORPH is shown in Fig. 1.
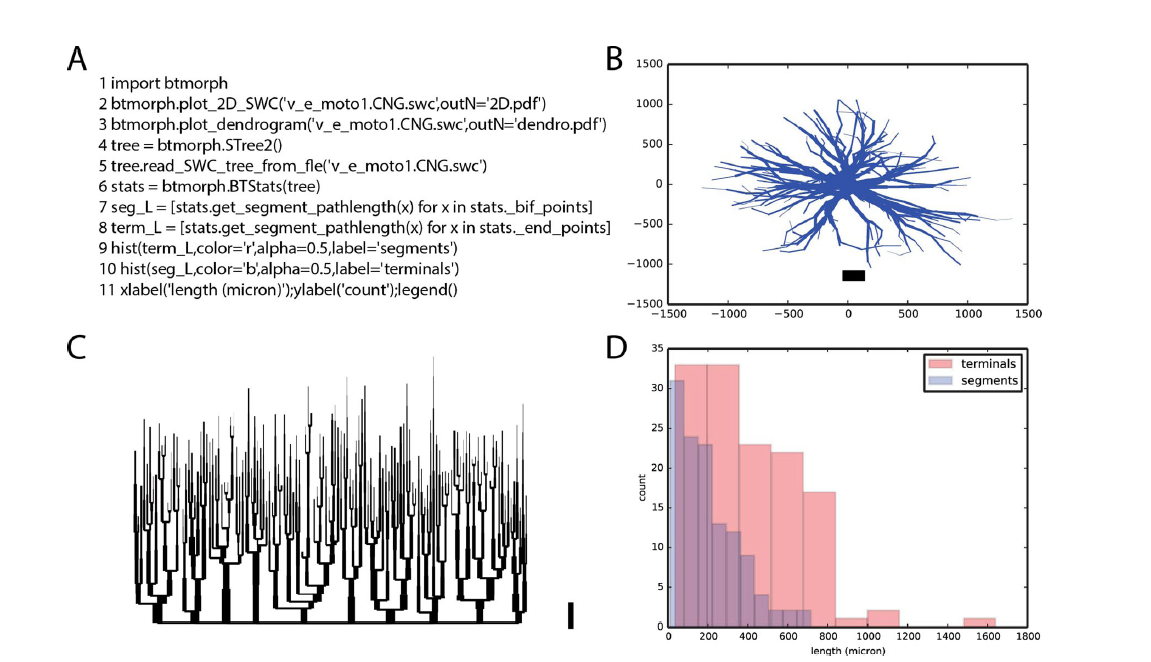
Figure 1: Exemplar use of BTMORPH A. Code snippet (line numbers added for clarity). B-D Results of executing the code snippet. B. line 2 (of the snippet) generates a 2D rendering of the morphology and save the file as PDF. C. line 3 generates a dendrogram of the same morphology. D. execution of lines 4–11 load a morphology into the provided datastructure (STree2) and computes and visualizes the segment length statistics of this neuron
Software for modeling growth of dendrites and axons
Neuronal morphologies are essential for brain function: physical overlap between dendrites and axons constrain the circuit topology, and the precise shape and composition of dendrites determine the integration of inputs to produce an output signal. At the same time, morphologies are highly diverse and variant. The variance, presumably, originates from neurons developing in a densely packed brain substrate where they interact (e.g., repulsion or attraction) with other actors in this substrate. However, when studying neurons their context is never part of the analysis and they are treated as if they existed in isolation. To fully understand neuronal morphology and its variance it is important to consider neurons in relation to each other and to other actors in the surrounding brain substrate, i.e., their context.
We developed a context-aware computational framework, NeuroMaC, in which large numbers of neurons can be grown simultaneously according to growth rules expressed in terms of interactions between the developing neuron and the surrounding brain substrate (Torben-Nielsen & De Schutter, 2014). NeuroMaC can generate accurate virtual morphologies of distinct classes both in isolation and as part of neuronal forests. Accuracy is validated against population statistics of experimentally reconstructed morphologies. We showed that context-aware generation of neurons can explain characteristics of variation. Indeed, plausible variation is an inherent property of the morphologies generated by context-aware rules (Fig. 2).
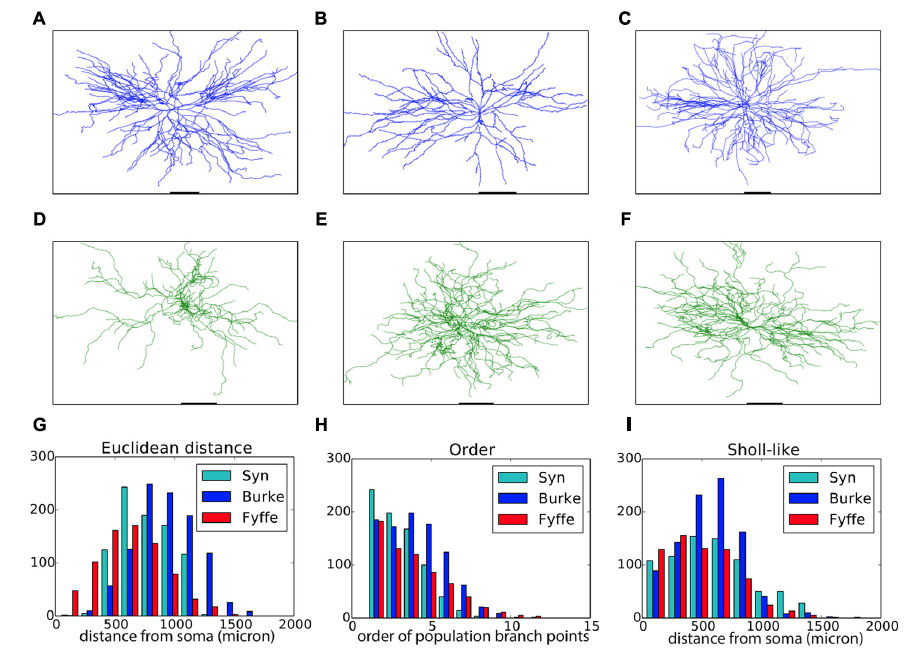
Figure 2: Validation of generated alpha motor neurons. A–C. Exemplar experimentally reconstructed spinal cord alpha motor neurons [A,B from the Fyffe archive (Alvarez etal., 1998), C from the Burke archive (Cullheim et al., 1987)]. D–F. Virtual morphologies generated by NeuroMaC. G–I. Quantitative comparison to archives mentioned, "Syn" are generated morphologies.
A demonstration of the effect of context on neural development is given in Fig. 3 which shows the generation of layer 5 pyramidal neuron morphologies. Three exemplar morphologies are shown in Fig. 3A. We can observe morphological traits such as a difference in “height” between the cells but these traits are hard to relate to their context. However, from canonical circuit information, we know that the somas are located in layer 5 (L5), that their basal dendrites remain mainly in L5 and may extend a bit into layer 4, that their apical dendrite extends to the superficial parts and ends close to the pia (in layer 1) after branching extensively in layers 1-3. The remarkable difference in “height” of the apical tree, is a clear signature of this context dependence as pyramidal cells more superficially located in L5 cannot extend as far as more deeply positioned ones. Ther generated morphologies clearly capture this behavior (Fig. 3B).
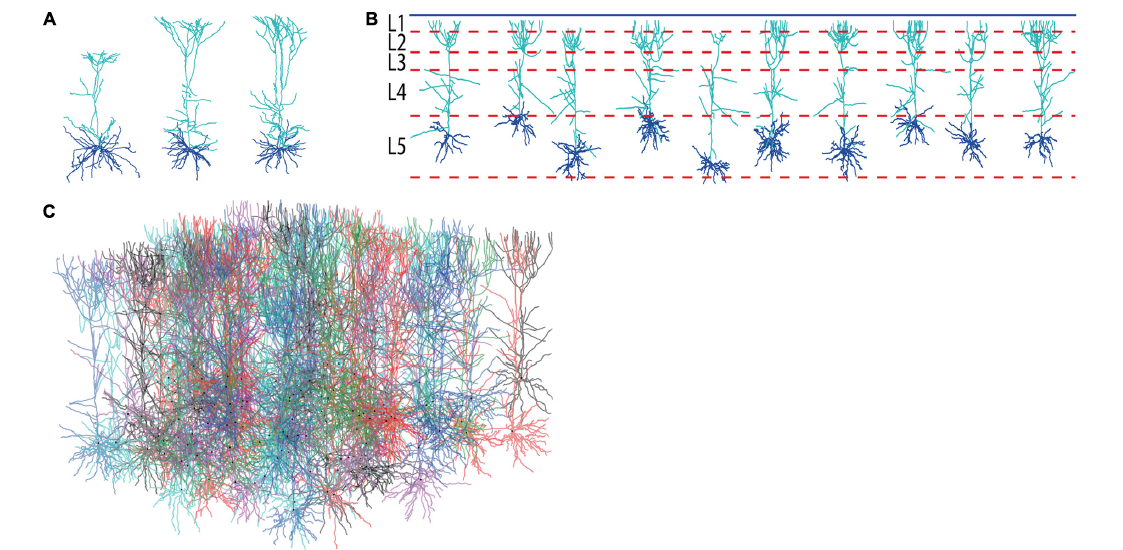
Figure 3: Generation of layer 5 pyamidal neurons. A. Experimentally reconstructed layer 5 pyramidal neurons (from the Kawaguchi archive, Hirai et al. 2012). B. Virtual morphologies generated by NeuroMaC. Simulated laminar structure (L1–L5, from top to bottom) indicated by dashed lines; blue line represents the pia. C. Forest of 100 simultaneously generated, non-overlapping pyramidal neurons.
Effect of morphology on intracellular calcium dynamics
There is growing interest in understanding calcium dynamics in dendrites, both experimentally and computationally. Many processes influence these dynamics, but in dendrites there is a strong contribution of morphology because the peak calcium levels are strongly determined by the surface to volume ratio (SVR) of each branch, which is inversely related to branch diameter. We explored the predicted variance of dendritic calcium concentrations due to local changes in dendrite diameter and how this is affected by the modeling approach used. We investigated this in a model of dendritic calcium spiking in different reconstructions of cerebellar Purkinje cells and in morphological analysis of neocortical and hippocampal pyramidal neurons (Anwar et al., 2014).
Traditionally, a Ca2+ pool with a single relaxation time constant is used to model intracellular Ca2+ dynamics. Such models compute the effects of Ca2+ influx accurately but combine all removal systems, including diffusion, into one process with a fixed time constant. They usually represent the Ca2+ concentration in a submembrane shell with a fixed depth. Previously, we have shown that these pool based models can not capture the complex dynamics of intracellular Ca2+ because they fail to simulate the multiple time scales at which interactions between Ca2+ channels and Ca2+-activated K+ channels occur (Anwar et al., 2012). In Anwar et al. (2014) we extended the comparison of Ca2+ pool to complex Ca2+ dynamics models to the spatial domain. We showed that many model implementations in the literature do not compute correct volumes for the submembrane shell, resulting in diameter-independent calcium concentrations in the model (Fig. 4A) and describe how to implement this correctly in the NEURON simulator, both for phenomenological pool based models and for implementations using radial 1D diffusion.
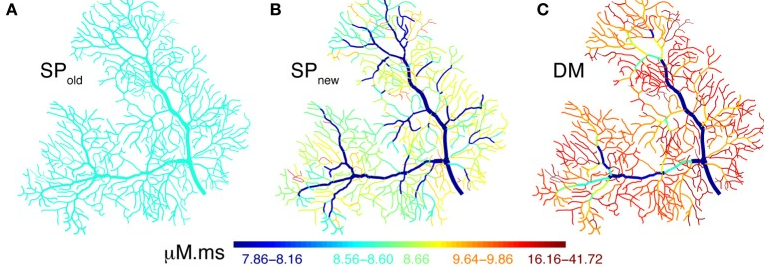
Figure 4: Spatial Ca2+ gradients strongly depend on type of model implementation. Panels A–C show maps of the integrated calcium levels in the dendrite during a spontaneous burst of dendritic Ca2+ spikes. The dendritic branches are color coded to show the integrated calcium levels using a 20 ms window around the peak Ca2+ concentration of the first Ca2+ spike. The color scales used in these maps are nonlinear (using histogram equalization) to enhance the contrast. A. Single Ca2+ pool model using a common error in implementation in the NEURON simulator results in homogenous Ca2+ levels. B. Single Ca2+ pool model using a corected NEURON mechanism results in variable Ca2+ levels. C. Detailed Ca2+ dynamics model with buffering and 1D diffusion results in variable Ca2+ levels with larger Ca2+ gradients.
But, even with a correct implementation of the submembrane shell, pool models predict substantially smaller Ca2+ concentrations than mode detailed models simulating buffered 1D diffusion (Fig. 4C). Are the latter models sufficient to predict neural calcium concentrations at the typical resolution of compartmental models or is full 3D diffusion required? This is investigated in Fig. 5 by comparing 1D diffusion models in the NEURON simulator with full 3D diffusion in the STEPS simulator. Detailed comparison shows that 1D diffusion of models of calcium buffering give a good approximation provided an accurate morphological reconstruction is available and modeled in high resolution (Fig. 5D).
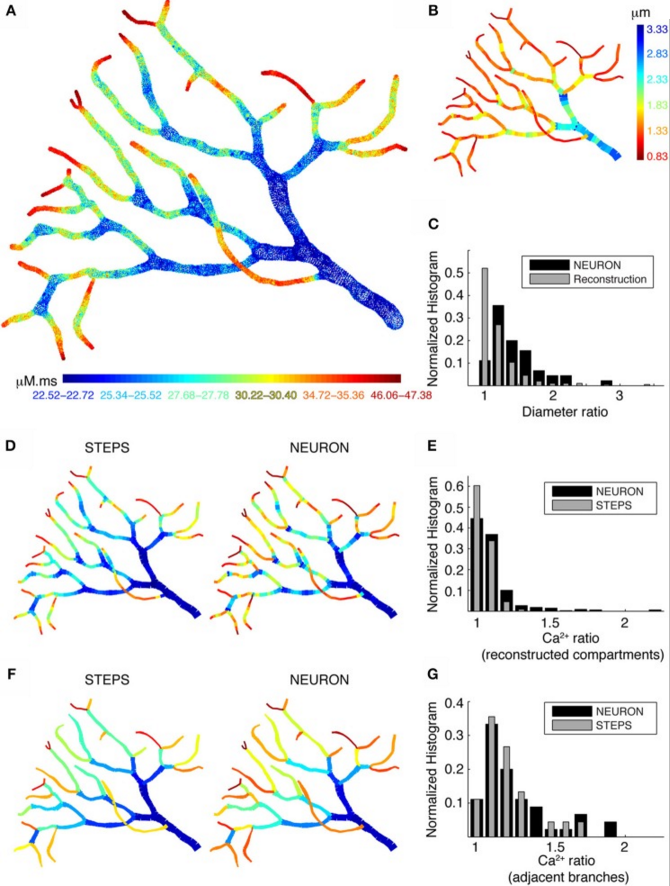
Figure 5: Large differences in calcium levels in adjacent dendritic branches persist in presence of 3D diffusion. A. STEPS model using 3D buffered diffusion to compute the Ca2+ concentration resulting from the burst of Ca2+ spikes. Spatial map of integrated calcium (140ms window) in a piece of carefully reconstructed PC dendritic arbor. B. Spatial map of dendritic diameters in the dendrite. C. Normalized histograms compare the ratios of adjacent diameters in the original morphological reconstruction with similar ratios of diameters of adjacent compartments in the NEURON model (1 segment per unbranched section), demonstrating a large difference between these two distributions. D, E NEURON simulation with many compartments for each unbranched segment, carefully reflecting the variability of dendrite diameter. Data for the STEPS simulation are averaged over all tetrahedrons representing the corresponding NEURON compartment. D shows the respective spatial maps which are similar to the reference case shown in A. E shows a normalized histograms of the ratios of integrated Ca2+ concentration between every adjacent compartment, demonstrating a reasonable match between the results from the two simulators. F,G NEURON simulation with a single compartment for each unbranched segment (a 'poor' reconstruction), data for the STEPS simulation averaged for corresponding NEURON compartments. Again the normalized histogram (G) shows a good match between the two simulators, but the spatial maps (F) are very different from the reference case (A).
4. Publications
4.1 Journals
- Anwar, H., Roome, J. C., Nedelescu, H., Chen, W., Kuhn, B. & De Schutter, E. Dendritic diameters affect the spatial variability of intracellular calcium dynamics in computer models. Frontiers in Cellular Neuroscience 8, 168, doi:10.3389 (2014).
- Couto, J., Linaro, D., De Schutter, E. & Giugliano, M. On the firing rate dependency of the phase response curve of rat Purkinje neurons in vitro. PLoS Computational Biology 11, e1004112 (2015).
- Laudanski, J., Torben-Nielsen, B., Segev, I. & Shamma, S. Spatially distributed dendritic resonance selectively filters synaptic input. PLoS Computational Biology 10, e1003775, doi:10.1371/journal.pcbi.1003775 (2014).
- Negrello M.: Valentino Braitenberg: From neuroanatomy to behavior and back. Biological Cybernetics 108: 527-539 (2014).
- Torben-Nielsen, B. An Efficient and Extendable Python Library to Analyze Neuronal Morphologies. Neuroinformatics 12, 619-622, doi: 10.1007/s12021-014-9232-7 (2014).
- Torben-Nielsen, B. & De Schutter, E. Context-aware modeling of neuronal morphologies. Frontiers in Neuroanatomy 8, 92, doi:10.3389/fnana.2014.00092 (2014).
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
Oral Presentations
- Hong, S. Patterns in network activity and information processing in a detailed computer model of the cerebellar granular layer, in CNS meeting, Quebec, Canada (2014).
- Hong, S. Multiplexed coding by cerebellar Purkinje neurons, in Recent Findings on the Cerebellar Microcircuitry, OIST Campus (2015).
- Torben-Nielsen, B. Purkinje cells: does the forest shape the trees?, in Recent Findings on the Cerebellar Microcircuitry, OIST Campus (2015).
- Close, T. Code generation from NineML: targeting NEURON and NEST simulators, Code Generation from Model Description Languages, Paris, France (2014).
- De Schutter, E. Stochastic effects of channel gating on neural excitability, Brain Corporation, San Diego, USA (2014).
- De Schutter, E. Stochasticity of Biological Synapses, NII Shonan Meeting, Kanagawa, Japan (2014).
- De Schutter, E. Importance of stochasticity and small molecule number in the induction of synaptic plasticity, Leiden, Germany (2014).
- De Schutter, E. Data publication to improve data sharing, Frankfurt Institute for Advanced Studies, Germany (2014).
- De Schutter, E. Parallelization of the spatial SSA on unstructured meshes, Banff, Canada (2014).
- De Schutter, E. Stochastic processes in Purkinje cell dendrites and spines, Institut des Neurosciences Cellulaires et Integratives, Strasbourg, France (2015).
- Hong, S. Exploring microcircuits in the cerebellum via computational modeling, Sungshin Women's University (Woonjung Campus), Seoul, Korea (2014).
- Torben-Nielsen, B. Context-aware modeling of neuronal morphologies & circuits, McGill University, Montreal, Canada (2014).
- Torben-Nielsen, B. Context-aware modeling of neuronal morphology, the Hebrew University, Jerusalem, Israel (2014).
- Torben-Nielsen, B. Context-aware modelling of neuronal morphologies and circuits, Riken BSI, Japan (2015).
- Torben-Nielsen, B. Purkinje cells: does the forest shape the trees?, Kyoto University, Japan (2015).
Poster Presentations
- Close, T. G., Torben-Nielsen, B. & De Schutter, E. Reduction of multi-compartmental biophysical models by incremental, automated retuning of their parameters and synaptic weights, in CNS meeting, Quebec, Canada (2014).
- Hepburn, I., Chen, W. & De Schutter, E. Accurate approximation and MPI parallelization of spatial stochastic reaction-diffusion in STEPS, in CNS meeting, Quebec, Canada (2014).
- Hong, S., Negrello, M., Junker, M., Smilgin, A., Thier, P. & De Schutter, E. Multiplexed coding by cerebellar Purkinje neurons, in Society for Neuroscience 2014, Washington DC, USA (2014).
- Raikov, I. & De Schutter, E. A computational exploration of connectivity in the cerebellar granular layer, in FENS 2014, Milan, Italy (2014).
- Raikov, I. & De Schutter, E. A NineML-based domain-specific language for computational in CNS Meeting, Quebec, Canada (2014).
- Rasumov, N. & De Schutter, E. ATP consumption in mATP consumption in molecular reactions of neuronal signalingolecular reactions of neuronal signaling, in CNS Unit, Quebec, Canada (2014).
- Sudhakar, S. K., Hong, S. & De Schutter, E. A computational study on the network dynamics and synchronization of cerebellar granular layer to different input patterns., in Society for Neuroscience meeting 2014, Walter E washington convention center, Washington DC, USA (2014).
- Torben-Nielsen, B. & De Schutter, E. A computational framework to study the influence of microscopic interactions on neuronal morphology, in CNS meeting, Quebec city, Canada (2014).
- Torben-Nielsen, B. & De Schutter, E. Context-aware modeling of neuronal morphology, in Society for Neuroscience 2014, Washington DC, USA (2014).
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 Summer Course
OIST Computational Neuroscience Course 2014
- Date: June 16 - July 3, 2014
- Venue: Seaside House, OIST
- Co-organizers: E. De Schutter, K. Doya & J. Wickens, OIST
- Co-sponsors: the European Commission, Seventh Framework Programme via the European project CSNII (FP7-ICT-601167)
- Speakers:
- Gordon Arbuthnott (OIST)
- Upinder Bhalla (NCBS, India)
- Claudia Clopath (Imperial College London, UK)
- Erik De Schutter (OIST)
- Kenji Doya (OIST)
- Jason Kerr (MPG Bonn, Germany)
- Bernd Kuhn (OIST)
- Javier Medina (University of Pennsylvania, USA)
- Hiroyuki Nakahara (RIKEN BSI, Japan)
- Yael Niv (Princeton University, USA)
- Tony Prescott (University of Sheffield, UK)
- Ivan Soltesz (UC Irvine, USA)
- Greg Stephens (OIST)
- Greg Stuart (Eccles Institute of Neuroscience, Australia)
- Jeff Wickens (OIST)
- Taro Toyoizumi (RIKEN BSI, Japan)
6.2 Mini Symposium
Recent Findings on the Cerebellar Microcircuitry
- Date: Jan 28 - 29, 2015
- Venue: OIST Main Campus (C210 Seminar Room, Center Building)
- Organizer : Erik De Schutter
- Speakers:
- Chris De Zeeuw (Netherlands Institute for Neuroscience)
- Stéphane Dieudonné (École Normale Supérieure)
- Michael Häusser (University College London)
- Sungho Hong (OIST)
- Masanobu Kano (University of Tokyo)
- Mineko Kengaku (University of Kyoto)
- Arthur Konnerth (Technische Universität München)
- Angus Silver (University College London)
- Keiko Tanaka-Yamamoto (Korea Institute of Science and Technology)
- Benjamin Torben-Nielsen (OIST)
- Marylka Yoe Uusisaari (Hebrew University of Jerusalem)
- Yosef Yarom (Hebrew University of Jerusalem)
6.3 Seminars
Title: Synaptic and non-synaptic learning correlates in neural circuits
- Date: Aug.18, 2014
- Venue: C015, Lab1, OIST
- Speaker : Prof. Dr. Christian Hansel - Neurobiology of learning & memory, the University of Chicago
Title: Dendritic cells for immunity and tolerance
- Date: Dec.1, 2014
- Venue: C015, Lab1, OIST
- Speaker: Prof. Zwi N. Berneman, MD PhD FRCP - Antwerp University Hospital and University of Antwerp, Antwerp, Belgium
7. Other
Nothing to report.



