FY2013 Annual Report
Computational Neuroscience Unit
Professor Erik De Schutter
Abstract
We use computational, data-driven methods to study how neurons and microcircuits in the brain operate. We are interested in how fundamental properties, such as a neuron’s morphology and its excitability, interact with one another during common neural functions like information processing or learning. Most of our models concern the cerebellum as this brain structure has a relatively simple anatomy and the physiology of its main neurons has been studied extensively, allowing for detailed modeling at many different levels of complexity.
1. Staff
General services and neuroinformatics
- Ivan Raikov, Technical Staff
- Tsuyuki Nakabayashi, Research Administrator/ Secretary (till May 2013)
- Sachie Matsuoka, Research Administrator/ Secretary
Molecular modeling
- Weiliang Chen, Researcher
- Iain Hepburn, Technical Staff
- Anant Jain, Technical Staff (till August 2013)
- Nikon Rasumov, Researcher (from May 2013)
Cellular modeling
- Sungho Hong, Group Leader
- Shiwei Huang, Researcher (till November 2013)
- Akira Takashima, Researcher
- Haroon Anwar, Technical Staff (till Octover 2013)
- Hermina Nedelescu, Special Research Student
Network modeling
- Benjamin Torben-Nielsen (from October 2013)
- Tom Close, Researcher
- Nol Chindapol, Researcher (from April till September 2013)
- Shyam Kumar Sudhakar, Special Research Student
Rotation students
- Chen Jiabao, Rotation Student (2nd term 2013)
- Igarashi Masakazu, Rotation Student (2nd term 2014)
2. Collaborations
- Theme: Cerebellar physiology, multiple themes
- Type of collaboration: Scientific collaboration and graduate program
- Researchers:
- Professor M. Giugliano, University of Antwerp, Belgium
- Professor D. Snyders, University of Antwerp, Belgium
- Joao Couto, University of Antwerp, Belgium
- Q. Robberecht, University of Antwerp, Belgium
- K. Veys, University of Antwerp, Belgium
- Theme: Spiking activity of monkey cerebellar neurons
- Type of collaboration: Scientific collaboration
- Researchers:
- Professor H.P. Thier, University of Tübingen, Germany
- A. Ignashchenkova, University of Tübingen, Germany
- Dr. M. Junker, University of Tübingen, Germany
- A. Schmigdlin, University of Tübingen, Germany
- Theme: Human Brain Project: simulator development
- Type of collaboration: Scientific collaboration
- Researchers:
- Prof. F. Schürmann, École Polytechnique Fédérale de Lausanne, Switzerland
- F. Delalondre, École Polytechnique Fédérale de Lausanne, Switzerland
- Theme: Molecular identification of cerebellar signaling pathways and cerebellar optogenetics
- Type of collaboration: Scientific collaboration
- Researchers:
- Professor K. Tanaka, Korea Institute for Science and Technology (KIST), Korea
- Professor K. Tanaka, Korea Institute for Science and Technology (KIST), Korea
- Theme: Modeling of effects of ethanol on the cerebellum
- Type of collaboration: Joint research
- Researchers:
- Professor C.F. Valenzuela, University of New Mexico, United States of America
- Professor C.F. Valenzuela, University of New Mexico, United States of America
- Theme: Correlation of neurons linked to their excitability
- Type of collaboration: Scientific collaboration
- Researchers:
- Professor S.A. Prescott, University of Toronto, Canada
- Dr. S. Ratté, University of Toronto, Canada
- Theme: Purkinje cell morphology and physiology, modeling
- Type of collaboration: Scientific collaboration
- Researchers:
- Professor M. Häusser, University College London, United Kingdom
- Professor H. Cuntz, Goethe University Germany
- Professor A. Watt, McGill University, Canada
- Dr. A. Roth, University College London, United Kingdom
3. Activities and Findings
3.1 Neuroinformatics standards
The work during the previous years on the NineML standard led to the submission of a paper (Raikov et al.) that is in the final stages of review. A NineML Standardization Committee was formed with Prof. De Schutter as acting chair.
3.2 Molecular mechanisms of synaptic plasticity
STEPS software development
STEPS is a stochastic reaction-diffusion simulation engine that implements a spatial extension of Gillespie's Stochastic Simulation Algorithm (SSA) in complex tetrahedral geometries. An extensive Python-based interface is provided to STEPS so that it can interact with the large number of scientific packages in Python. STEPS version 2.0 was released which includes accurate and efficient computation of local membrane potentials on tetrahedral meshes, with the addition of voltage-gated channels and currents. This is based on a novel method for calculating the quasi-static electrical potential on tetrahedral meshes, called E-Field (Hepburn et al. 2013). The application of E-Field on the same tetrahedral mesh provides a level of integration between electrical excitability and spatial molecular dynamics in realistic cellular morphology not previously achievable. Deterministic solutions are also possible. By performing the Rallpack tests we demonstrated the accuracy of the E-Field method. Efficient node ordering is an important practical consideration, and we find that a breadth-first search provides the best solutions, although principal axis ordering suffices for some geometries (Figure 1). The E-Field implementation in STEPS will play an important role in the future of multiscale neural simulations.
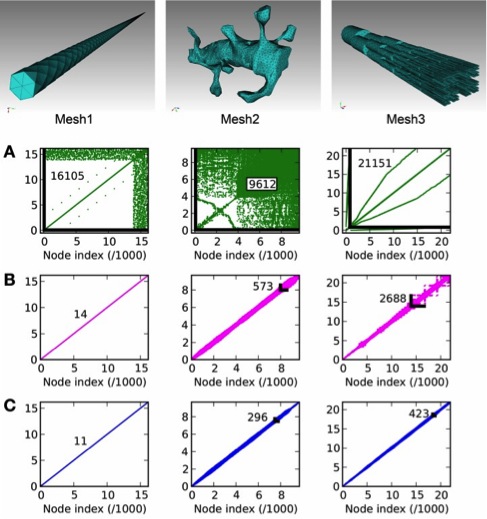
Figure 1: Node connectivity and its effect on E-Field calculation period. Connectivity matrices are
displayed for three tetrahedral meshes of different geometries, with A: no ordering (mesh generator
output), B: principal axis ordering, C: breadth-first search ordering. The maximum node separation
is displayed for each connectivity matrix as a number and, where visible, a black line showing the
maximally separated node connection.
The tight integration between the reaction-diffusion calculations and the tetrahedral mesh potentials allows detailed coupling between molecular activity and local electrical excitability. A later STEPS update also included two new supporting toolkits that support geometry preparation and visualization for STEPS simulations (Chen and De Schutter 2014) (Figure 2).
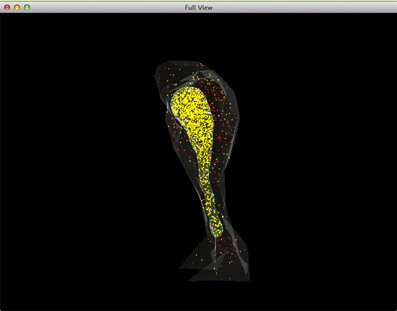
Figure 2: A complete view of a STEPS simulation of Ca2+ release by IP3 receptors on the endoplasmatic
reticulum (ER). Red: IP3 in cytosol, Orange: Ca2+ (Ca2+ in ER is rendered as yellow due to the color
combination with ER compartment), Blue: inactivated IP3 receptors, Dark magenta: IP3 bound
receptors, Bright magenta: IP3 receptors in open state.
The role of dendritic spine morphology in compartmentalizing surface receptors
Since AMPA receptors are major molecular players in both short- and long-term plasticity, it is important to identify the time-scales of and factors affecting the lateral diffusion of AMPARs on the dendrite surface. We studied how dendritic spine morphology affects two processes: (1) compartmentalization of the surface receptors in a single spine to retain local chemistry and (2) the delivery of receptors to the post-synaptic density (PSD) of spines via lateral diffusion following insertion onto the dendrite shaft (Simon et al. 2013). Computing the mean first passage time (MFPT) of surface receptors on a EM reconstructions of of real spines revealed that a constricted neck and bulbous head serve to compartmentalize receptors. We found that the location of the PSD corresponds to the location at which the maximum MFPT occurs, the position that maximizes the residence time of a diffusing receptor (Figure 3).
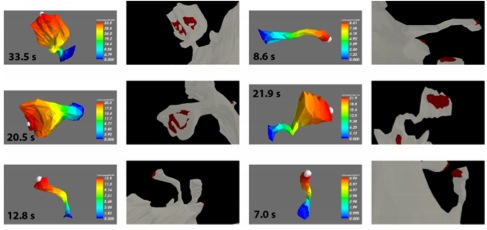
Figure 3: The mean first passage time (MFPT), a measure of how fast receptors diffuse away, as a
function of the initial position the spine, next to the image of the spine on the dendrite with the
excitatory postsynaptic density indicated in red. The maximum of the MFPT is indicated by the
white sphere for comparison to the location of the synaptic region. Maximum MFPT is written
in black text.
Meanwhile, the same geometric features of the spine that compartmentalize receptors inhibit the recruitment of AMPARs via lateral diffusion from dendrite insertion sites. Spines with narrow necks will trap a smaller fraction of diffusing receptors in the their PSD when considering competition for receptors between the spines (Figure 4), suggesting that ideal geometrical features involve a tradeoff depending on the intent of compartmentalizing the current receptor pool or recruiting new AMPARs in the PSD. The ultimate distribution of receptors among the spine PSDs by lateral diffusion from the dendrite shaft is an interplay between the insertion location and the shape and locations of both the spines and their PSDs.
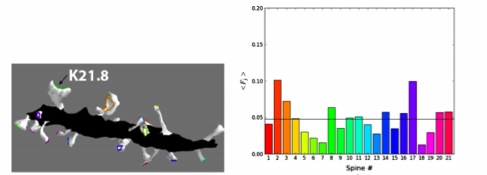
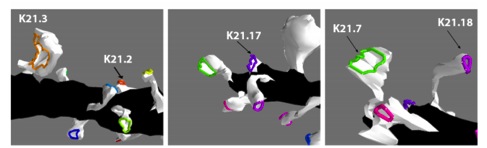
Figure 4: How spine shape and PSD area affect the probability of diffusing receptors reach the
postsymaptic density with uniform insertion of receptors along the dendrite shaft (black surface).
Top left: the distribution ofhe receptors among the spines deviates substantially from a uniform
distribution, revealing that spine/PSD geometry plays a large role in receptor recruitment. Other
panels: labeling and shape of some of the spines.
3.3 Cellular mechanisms regulating firing and synaptic properties of neurons
Stochastic dendritic calcium spikes
Bursts of dendritic calcium spikes play an important role in excitability and synaptic plasticity in many types of neurons. In single Purkinje cells, spontaneous and synaptically evoked dendritic calcium bursts come in a variety of shapes with a variable number of spikes. The mechanisms causing this variability have never been investigated thoroughly.We used a detailed computational model using novel simulation routines to identify the roles that stochastic ion channels, spatial arrangements of ion channels, and stochastic intracellular calcium have toward producing calcium burst variability (Anwar et al. 2013). Consistent with experimental recordings from rats, strong variability in the burst shape is observed in simulations. This variability persists in large model sizes in contrast to models containing only voltage-gated channels, where variability reduces quickly with increase of system size. Phase plane analysis of Hodgkin-Huxley spikes and of calcium bursts identifies fluctuation in phase space around probabilistic phase boundaries as the mechanism determining the dependence of variability on model size. Stochastic calcium dynamics are the main cause of calcium burst fluctuations, specifically the calcium activation of mslo/BK-type and SK2 channels (Figure 5). Local variability of calcium concentration has a significant effect at larger model sizes.
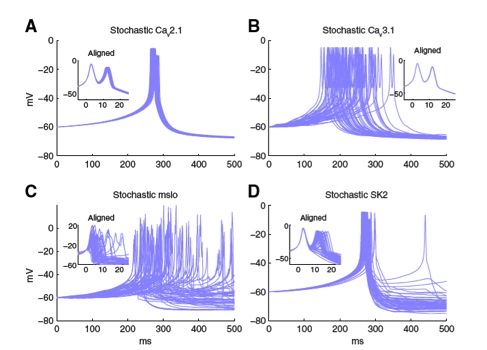
Figure 5: The stochastic effect of Ca2+-activated K+ channels (C, D) is much larger than that of voltage-gated
Ca2+ channels (A, B), causing a large variability in calcium burst shape and jitter of spike timing. STEPS
stimulation of a hybrid model of a calcium burst where only one channel type is simulated in stochastic mode,
all other mechanisms of the model including the other channels and calcium dynamics are simulated
deterministically.
Simulations of both spontaneous and synaptically evoked calcium bursts in a reconstructed dendrite show, in addition, strong spatial and temporal variability of voltage and calcium, depending on morphological properties of the dendrite (Figure 6). Our findings suggest that stochastic intracellular calcium mechanisms play a crucial role in dendritic calcium spike generation and are therefore an essential consideration in studies of neuronal excitability and plasticity.
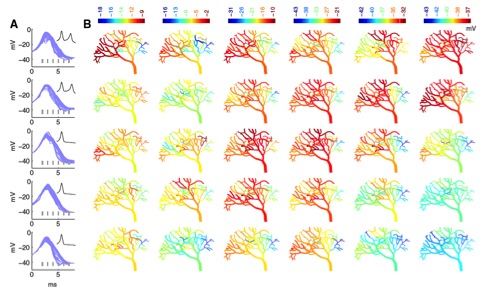
Figure 6: Fully stochastic calcium burst simulation in a dendritic branch shows large spartiotemporal variation
of membrane potentials. Each row represents a different run of the stochastic model of a Purkinje cell dendrite
with shapes of the bursts shown in A (full burst in inset, top two rows have two spikes in the burst, bottom ones
only one spike). B: Spatial maps of membrane potential at each time point marked with gray lines in A. Each
time point has its own color scale to emphasize differences within single maps.
3.4 Information processing in the olivocerebellar system
Multiplexing of neuronal codes
Neurons are often said to operate as integrators or as coincidence detectors based on how they process input. Integrators can summate temporally dispersed (asynchronous) inputs, whereas coincidence detec-tors respond only to temporally coincident (synchronous) inputs. In other words, integrators and coincidence detectors are both sensitive to synchronous input, but coincidence detectors are selective for it. The terms integration and coincidence detection serve to highlight differences in neuronal operation, but they do not accurately depict how an average neuron operates. Some neurons are exquisitely specialized to operate in one or the other mode but most, including the average pyramidal neuron, operate somewhere in between. In that respect, operating mode is best conceptualized not as a dichotomy, but rather as a continuum with ‘‘pure’’ integration and ‘‘pure’’ coincidence detection at either end (Figure 7). Neurons operating in the midrange may exhibit traits of both operating modes, with certain traits manifesting more strongly than others depending on stimulus properties. By not being optimized for integration or coincidence detection, neurons can exhibit traits of both operating modes and could, therefore, be said to use a hybrid mode. This raises the question of whether a hybrid operating mode conveys benefits that justify the lack of specialization. In Ratté et al. (2013) we propose that a hybrid operating mode allows rate and synchrony codes to be multiplexed. Multiplexing refers to the transmission of more than one signal via a single communication channel and can increase information capacity.
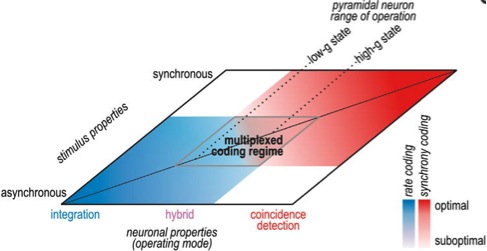
Figure 7: Neural coding depends jointly on neuronal operating mode and stimulus properties. Neuronal
operating mode is represented as a continuum on one axis. Pyramidal neurons tend to operate in the middle
range and can shift where they operate based on factors like conductance state. Input synchrony is
represented on the other axis. Neural coding strategies are represented in blue (rate coding) and red
(synchrony coding), and deeper colors represent better coding than paler colors. Pale regions overlap,
revealing a regime in which a hybrid operating mode and multiplexed coding are possible.
4. Publications
4.1 Journals
- Anwar, H., Hepburn, I., Nedelescu, H., Chen, W. & De Schutter, E. Stochastic calcium mechanisms cause dendritic calcium spike variability. Journal of Neuroscience 33, 15848-15867 (2013).
- Chen, W. & De Schutter, E. Python-based geometry preparation and simulation visualization toolkits for STEPS. Frontiers in Neuroinformatics 8, 37, doi:10.3389/fninf.2014.00037 (2014).
- De Schutter, E. Collaborative modeling in neuroscience: time to go open model? Neuroinformatics 11, 135-136, doi:10.1007/s12021-013-9181-6 (2013).
- De Schutter, E. The dangers of plug-and-play simulation using shared models. Neuroinformatics 12, 227-228 (2014).
- Hepburn, I., Cannon, R. & De Schutter, E. Efficient calculation of the quasi-static electrical potential on a tetrahedral mesh and its implementation in STEPS. Frontiers in Computational Neuroscience 7, 129 (2013).
- Huang, S. & Uusisaari, M. Y. Physiological temperature during brain slicing enhances the quality of acute slice preparations. Frontiers in cellular neuroscience 7, 48, doi:10.3389/fncel.2013.00048 (2013).
- Ratte, S., Hong, S., De Schutter, E. & Prescott, S. A. Impact of neuronal properties on network coding: roles of spike initiation dynamics and robust synchrony transfer. Neuron 78, 758-772, doi:10.1016/j.neuron.2013.05.030 (2013).
- Simon, C., Chen, W., Hepburn, I. & De Schutter, E. The role of dendritic spine morphology in the compartmentalization and delivery of surface receptors. Journal of Computational Neuroscience, doi:10.1007/s10827-013-0482-4 (2013).
- Veys, K., Snyders, D. J. & De Schutter, E. Kv3.3b expression defines the shape of the complex spike in the Purkinje cell. Frontiers in cellular neuroscience 7, 205 (2013).
4.2 Books and Other One-Time Publications
Nothing to report
4.3 Oral and Poster Presentations
Oral Presentations
- De Schutter, E. Data publication to improve data sharing, in Polish Neuroscience Society Congres, Poznan, Poland (2013).
- Close, T. G., Raikov, I. & De Schutter, E. A software framework for developing large-scale, biophysically constrained neuronal simulations, in NeuroEng 2014, the 7th Australian workshop on computational neuroscience, National Wine Centre, Adelaide, Australia (2014).
- De Schutter, E. Stochastic calcium spikes in Purkinje cell dendrites, in Computational Neuroscience Workshop, CNS 2013, Computations in the cerebellar circuit, Paris, France (2013).
- De Schutter, E. The effects of passive membrane non-linearity on neuro-excitability, in Computational Neuroscience Workshop, CNS 2013, Computational properties of inhibitory synapses, Paris, France (2013).
- Close, T. G. Towards a biophysically inspired model of the cerebellum, the Department of Electrical and Electronic Engineering at the University of Melbourne (2014).
- De Schutter, E. Coding in silence: cerebellar Purkinje cell pauses control eye saccade endings., The Ninth Computational Motor Control Workshop, Beer-Sheva, Israel (2013).
- De Schutter, E. Integrated membrane potential and reaction-diffusion modeling on submicron resolution meshes, INCF workshop: Methods in bridging subcellular and cellular level models, San Diego, USA (2013).
- De Schutter, E. Challenges in multi-scale modeling of the brain, Rusutsu, Hokkaido Japan (2014).
- Hong, S. Multiplexed coding of the cerebellar Purkinje neurons, De Zeeuw lab, Erasmus MC, Rotterdam, Netherlands (2013).
Poster Presentation
- Chen, W., Hepburn, I. & De Schutter, E. Dendritic Volume Mesh Reconstruction for STEPS: How Does Mesh Quality Affect Stochastic Reaction-Diffusion Simulation?, in CNS 2013 Paris, Paris, France (2013).
- Close, T. G., Raikov, I. & De Schutter, E. A multi-simulator framework for accessible large-scale simulations of biophysical neuronal networks., in The 34th Annual meeting of Australian Neuroscience Society Meeting, Adelaide Convention Centre, Adelaide, Australia (2014).
- Close, T. G., Raikov, I., Kumar, S. & De Schutter, E. Exploring the limitations of simulator independence via an implementation of a biophysically detailed cerebellar cortex model in NEURON and NEST in CNS 2013 Paris, Paris, France (2013).
- Hong, S., Negrello, M., Junker, M., Thier, P. & De Schutter, E. Saccade angle modulates correlation between the local field potential and cerebellar Purkinje neuron activity, in CNS 2013 Paris, Paris, France (2013).
- Jain, A., Hepburn, I., Chen, W. & De Schutter, E. Raf-1 sensitivity to MAPK positive feedback loop maintaining long term depression in purkinje cells, in CNS 2013 Paris, Paris, France (2013).
- Kumar, S. & De Schutter, E. A biophysical model of cerebellar molecular layer interneuron, in CNS 2013 Paris, Paris, France (2013).
- Raikov, I., Close, T. G., Kumar, S. & De Schutter, E. Challenges of declarative modeling of conductance-based neurons in diverse simulation environments in CNS 2013 Paris, Paris, France (2013).
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 Summer Course
OIST Computational Neuroscience Course 2013
- Date: June 17- July 4, 2013
- Venue: Seaside House, OIST
- Co-organizers: E. De Schutter, K. Doya & J. Wickens, OIST
- Co-sponsors: the European Commission, Seventh Framework Programme via the European project CSNII (FP7-ICT-601167)
- Speakers:
- Angelo Arleo (Université Pierre & Marie Curie, France)
- Avrama Blackwell (George Mason University, USA)
- Erik De Schutter (OIST)
- Karl Deisseroth (Stanford University, USA)
- Sophie Deneve (Ecole Normale Supérieure, France)
- Kenji Doya (OIST)
- Gaute Einevoll (Norwegian University of Life Sciences)
- Mike Hasselmo (Boston University, USA)
- William Holmes (Ohio University, USA)
- Mitsuo Kawato (ATR, Japan)
- Bernd Kuhn (OIST)
- Jonathan Pillow (University Texas Austin, USA)
- Greg Stephens (OIST)
- Jeff Wickens (OIST)
- Yoko Yazaki-Sugiyama (OIST)
6.2 Winter Course
Course in Molecular Neuroanatomy 2014
- Date: January 27- February 2, 2014
- Venue: Seaside House, OIST
- Co-organizers: E. De Schutter, OIST & T. Gilbert, AIBS
- Co-sponsors: The Allen Institute for Brain Science
- Speakers:
- Gordon Arbuthnott (OIST)
- Erik De Schutter (OIST)
- David Feng (AIBS)
- Terri Gilbert (AIBS)
- Luis Puelles (University of Murcia, Spain)
- Josh Royall (AIBS)
- Izumi Sugihara (Tokyo Medical and Dental University, Japan)
- Charles Watson (Curtin University, Australia)
6.3 Seminars
Title: Expression and maintenance of cerebellar long-term synaptic depression
- Date: July 11, 2013
- Venue: C016, Lab1, OIST
- Speakers: Keiko Tanaka-Yamamoto, Ph.D.
-
Research Scientist at Center for Functional Connectomimcs, Korea Institute of Science and Technology
7. Others
Nothing to report



