FY2023 Annual Report
Computational Neuroscience Unit
Prof. Erik De Schutter

Front: Dr. Weiliang Chen, Ryo Nakatani, Chie Narai, Dr.Yinyun Li, Dr. Sungho Hong, Tomohiko Taniguchi (rotation student)
Abstract
We use computational, data-driven methods to study how neurons and microcircuits in the brain operate. We are interested in how fundamental properties, such as a neuron’s nanoscale morphology and its excitability, interact with one another during common neural functions like information processing or learning. Most of our models concern the cerebellum as this brain structure has a relatively simple anatomy and the physiology of its main neurons has been studied extensively, allowing for detailed modeling at many different levels of complexity. More recently we have also built models of hippocampal synapses.
1. Staff
- Molecular modeling
- Ryo Nakatani, PhD Student
- Sarah Yukie Nagasawa, PhD Student (till October 2023)
- Cellular modeling
- Sungho Hong, Group Leader (till March 2024)
- Gabriela Cirtala, Postdoctoral scholar
- Yinyun Li, Postdoctoral scholar (from November 2023)
- Information processing in the olivocerebellar system
- Juliana Silva de Deus, PhD Student (from April 2023)
- Software development
- Weiliang Chen, Staff Scientist
- Iain Hepburn, Technical Staff
- Jules Lallouette, Postdoctoral scholar
- Yifei Ma, Technical Staff (from January 2024)
- Visiting Researcher
- Andrew Gallimore, KAKENHI co-PI
- Christos Kotsalos, EPFL
- Tristan Carel, EPFL
- Taro Yasuhi,Yamaguchi University School of Medicine
- Rotation Students
- Tomohiko Taniguchi
- Research Interns
- Pin-Ju Chou
- Margaux Asclipe
- Research Unit Administrator
- Chie Narai
2. Collaborations
- Theme: Spiking activity of monkey cerebellar neurons
- Type of collaboration: Scientific collaboration
- Researchers:
- Professor H.P. Thier, University of Tübingen, Germany
- Akshay Markanday, University of Tübingen, Germany
- Junya Inoue, University of Tübingen, Germany
- Theme: Human Brain Project: simulator development
- Type of collaboration: Scientific collaboration
- Researchers:
- Prof. F. Schürmann, École Polytechnique Fédérale de Lausanne, Switzerland
- Dr. T. Carel, École Polytechnique Fédérale de Lausanne, Switzerland
- Dr. G. Castiglioni, École Polytechnique Fédérale de Lausanne, Switzerland
- Dr. A. Cattabiani, École Polytechnique Fédérale de Lausanne, Switzerland
- Theme: Quantitative molecular identification of hippocampal synapses
- Type of collaboration: Scientific collaboration
- Researchers:
- Prof. Dr. Silvio O. Rizzoli, Medical University Göttingen, Germany
- Dr. Svilen V. Georgiev Medical University Göttingen, Germany
- Theme: Cerebellar anatomy and physiology
- Type of collaboration: Scientific collaboration
- Researchers:
- Professor C. De Zeeuw, Erasmus Medical Center, Rotterdam, The Netherlands
- Professor L.W.J. Bosman, Erasmus Medical Center, Rotterdam, The Netherlands
- Dr. M. Negrello, Erasmus Medical Center, Rotterdam, The Netherlands
- Theme: Purkinje cell morphology and physiology, modeling
- Type of collaboration: Scientific collaboration
- Researchers:
- Professor M. Häusser, University College London, United Kingdom
- Professor A. Roth, University College London, United Kingdom
- Dr. S. Dieudonné, Ecole Normale Supérieure, Paris, France
- Taro Yasuhi,Yamaguchi University School of Medicine
- Theme: Circadadian rhythm generation
- Type of collaboration: Scientific collaboration
- Researchers:
- Professor J. Myung, Taipei Medical University, Taiwan
3. Activities and Findings
3.1 Software Development
NeuroDevSim: efficient simulation of neural development using shared memory parallelization
The Neural Development Simulator, NeuroDevSim, is a Python module that simulates brain development (De Schutter 2024): morphological growth, migration and pruning. It uses an agent-based modeling approach inherited from the NeuroMaC software (Torben-Nielsen and De Schutter 2014). Each cycle, agents called fronts execute model specific code. In the case of a growing dendritic or axonal front this will be a choice between extension, branching or growth termination. Somatic fronts can migrate to new positions and any front can be retracted to prune parts of neurons.
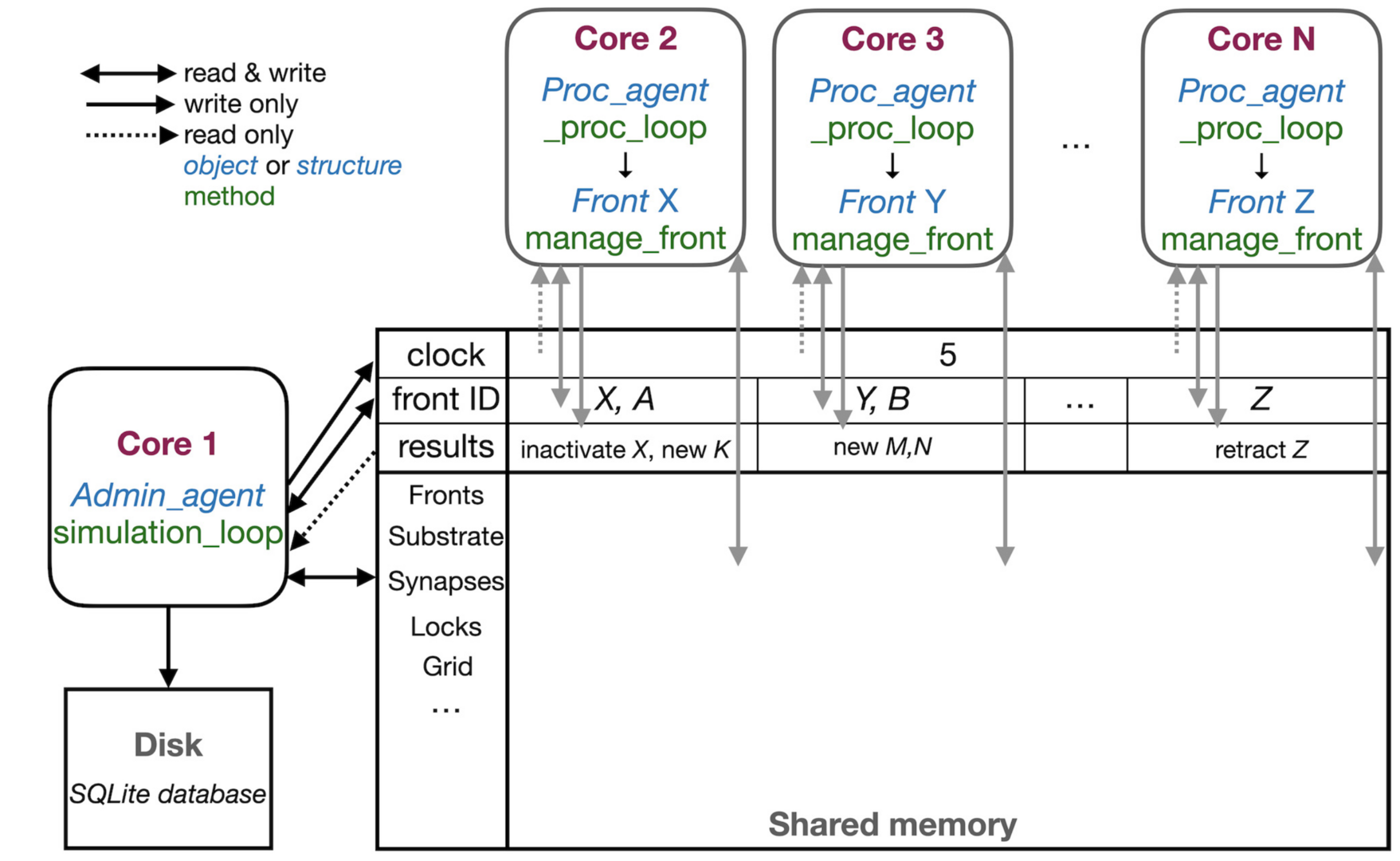
Figure 1: Schematic representation of the parallel workflow using shared memory. The simulation is controlled by Admin_agent that runs on Core 1 (left). Its interaction with processing cores during the execution of simulation_loop is shown. Admin_agent controls the clock that synchronizes all cores, which is available in shared memory. When a new simulation starts, Admin_agent puts the front IDs of fronts to be processed in dedicated areas of shared memory for each processing Core 2-N (top). The latter run _proc_loop that fetches the front corresponding to the ID executes its manage_front method and puts any new structures in the results array in shared memory as their ID plus a symbol. Admin_agent then fetches these results to store them in the database (bottom left) and to update its list of active fronts for the next cycle (from De Schutter 2023).
NeuroDevSim is a multi-core program that uses an innovative shared memory approach to achieve parallel processing without messaging (Figure 1). We demonstrated close to linear strong scaling for medium size models for up to 32 cores and have run large models successfully on 128 cores. Most of the shared memory parallelism is achieved without memory locking. Instead cores have only write privileges to private sections of arrays, while being able to read the entire shared array. Memory conflicts are avoided by a coding rule that allows only active fronts to use methods that need writing access. The exception is collision detection, needed to avoid growth of physically overlapping structures. Here a custom memory locking mechanism was necessary to control access to grid points that register the location of nearby fronts.
3.2 Network modeling
Model of early cerebellar development
We investigated the relationship between primary dendrite selection of Purkinje cells and migration of their presynaptic partner granule cells during early cerebellar development from P1 to P14. Through postnatal development, each Purkinje cell grows more than three dendritic trees, from which a primary tree is selected to develop further, whereas the others completely retract. Experimental studies suggest that this selection process is coordinated by physical and synaptic interactions with granule cells, which undergo a massive migration at the same time (Figure 2).
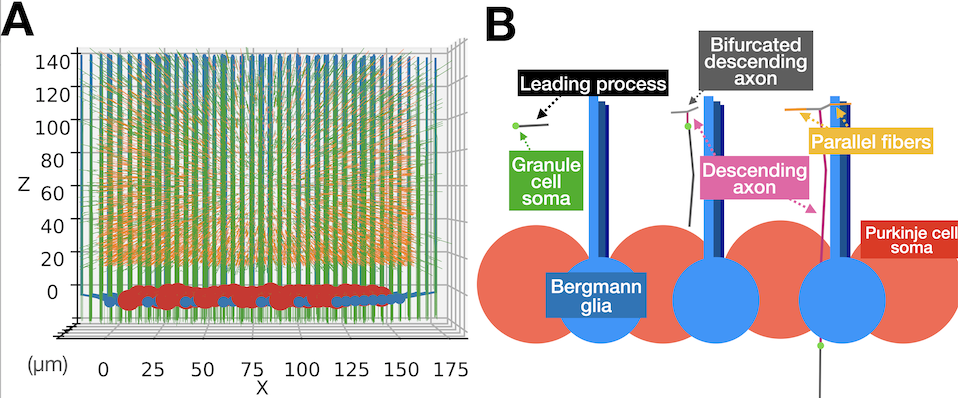
Figure 2: Granule cell migration model as an environment for Purkinje cell growth. (A) A representative simulation cube at the end of the simulation visualized from X side. Red large spheres are Purkinje cell somas. Blue small spheres with processes growing upward are Bergmann glia. Numerous green thin threads are axons of granule cells in the main simulation volume and orange fibers are ingrowing parallel fibers. (B) Schematic representations of how each granule cell migrates using a Bergmann glia process as a guide (from Kato and De Schutter 2023).
To explore possible mechanisms underlying this selection process, we constructed a computational model using the NeuroDevSim software (section 3.1). The study presents the first computational model that simultaneously simulates growing Purkinje cells and the dynamics of granule cell migrations during the first two postnatal weeks, allowing exploration of the role of physical and synaptic interactions upon dendritic selection. The model suggests that interaction with parallel fibers is important to establish the distinct planar morphology of Purkinje cell dendrites (Figure 3A). Specific rules to select which dendritic trees to keep or retract result in larger winner trees with more synaptic contacts than using random selection. A rule based on afferent synaptic activity was less effective than rules based on dendritic size or numbers of synapses (Figure 3B,C).

Figure 3: Resulting Purkinje cell dendritic trees after retraction of extra trees. (A) Examples of Purkinje cell trees: side and top views. (B,C) Retraction based on the sizes of different trees (B) results in significantly more parallel fiber synapses on the winner tree than retraction based on integrated synaptic input (C) (from Kato and De Schutter 2023).
3.3 Information processing in the olivocerebellar system
Multidimensional cerebellar computations
Both the environment and our body keep changing dynamically. Hence, ensuring movement precision requires adaptation to multiple demands occurring simultaneously. In this study we measured how monkey cerebellum adapts to general loss of motivation (“cognitive fatigue”) expressed as a reduction of the peak velocity of saccadic eye movements over 400 trials (Markanday, Hong et al. 2023). The reduction in eye velocity was compensated by a gradual upregulation of saccade duration (not shown) ensuring that endpoint accuracy was maintained. The cerebellum performed the necessary multi-dimensional computations for the flexible control of these movement parameters. This conclusion was based on the identification of a manifold-like activity in both mossy fibers (MFs, network input) and Purkinje cell simple spikes (PC-SS, output) (Figure 4). Unlike MFs, the PC manifolds developed selective representations of individual movement parameters. While the MF manifolds were characterized by an overall PV-related increase in their size almost symmetrically around the saccade onsets, the PC-SS manifolds on the first two dimensions showed no significant changes before saccade onsets, as depicted by the strong overlapping of the manifolds (Figure 4a-n). Instead pre-onset changes appeared only for the third and fourth dimensions of the PC manifolds (not shown). Furthermore, PC manifolds also carried a more disentangled representation of the two saccade parameters, peak velocity and duration, compared to MFs: in PCs peak eye velocity changed manifold size and saccade duration changed manifold rotation speed, while in MFs these effects were mixed (Figure 4p).
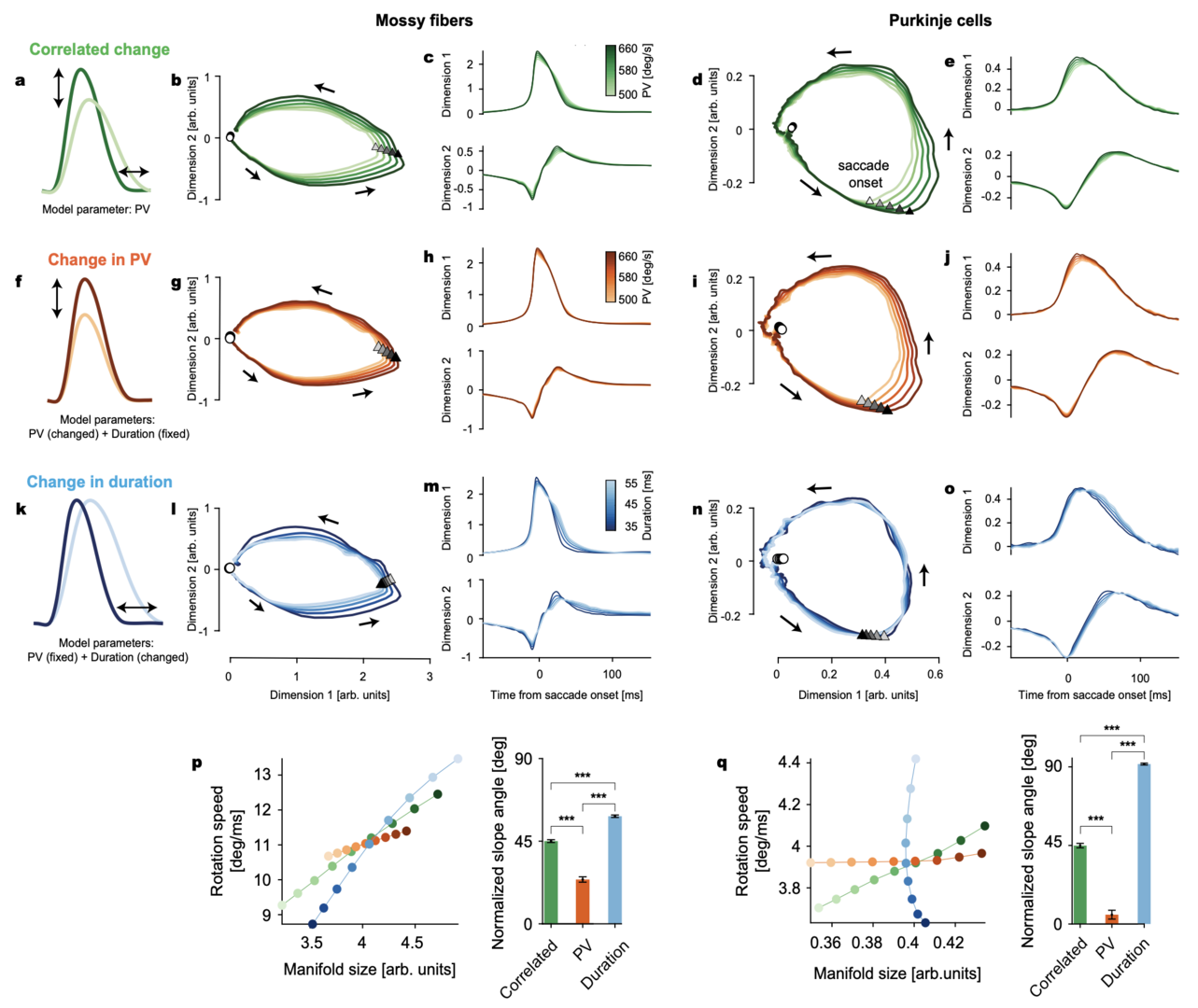
Figure 4: Manifolds identified in MF and PC-SS activity perform multi-dimensional encoding of eye movements. a Correlated changes in peak velocity (PV) and duration when PV is used as the only control parameter. b 2D plot of the first two dimensions in the MF manifold. Triangles and circles mark the saccade onsets and 250 ms before saccade onsets, respectively. Arrows show the direction of rotation. c The first two dimensions in b, plotted in time. d, e Same as b, c for PCs. f Isolated changes in saccade PV with the duration kept constant. g, h Isolated PV-dependent changes in the MF manifold computed from the rate models parametrized by PV but with fixed duration. i, j Same as g, h for PCs. k Isolated changes in saccade duration with constant PV. l–o Same as g, h and i, j but for duration change. p Left: MF manifold size versus rotation speed along the MF manifold varying with the correlated (green; a) and independent (orange and blue; f, k) change of PV and duration. Colors are as the color bars in c, h, m. Right: slope angle of the lines in left (from Markanday, Hong et al. 2023).
Error feedback-driven CF input modulated the PC manifolds to predict specific, error type-dependent changes in subsequent actions (not shown). Furthermore, a feed-forward network model that simulated MF-to-PC transformations revealed that amplification and restructuring of the lesser variability in the MF activity is a pivotal circuit mechanism (Figure 5). The number of dimensions that need to be considered to account for the properties of MF activity was much smaller than the maximum number of dimensions (116), but significantly higher than two or four, the dimensionalities capturing a major chunk of cell-to-cell variability in MFs and PCs, respectively (Figure 5). The PC-SS manifold (in the first two dimensions) was relatively poorly predicted (R2 < 0.9) when less than 9 MF dimensions were used (Figure 5f, g). Note that the dimensions higher than four explained only 4.3% of the total MF-to-MF variance together due to rapid decay in the explained variance. Therefore, the properties of multi-dimensional control of movements by PC-SS manifolds emerge from transformations by the cerebellar cortical circuit that amplify very small variabilities in MF inputs. This expansion of variability may allow PCs to extract as much information as possible during the movement so it can be used optimally when errors occur. Therefore, the cerebellar circuit can predict the dynamical, trial-to-trial deviations from the forward model for the average eye movement.
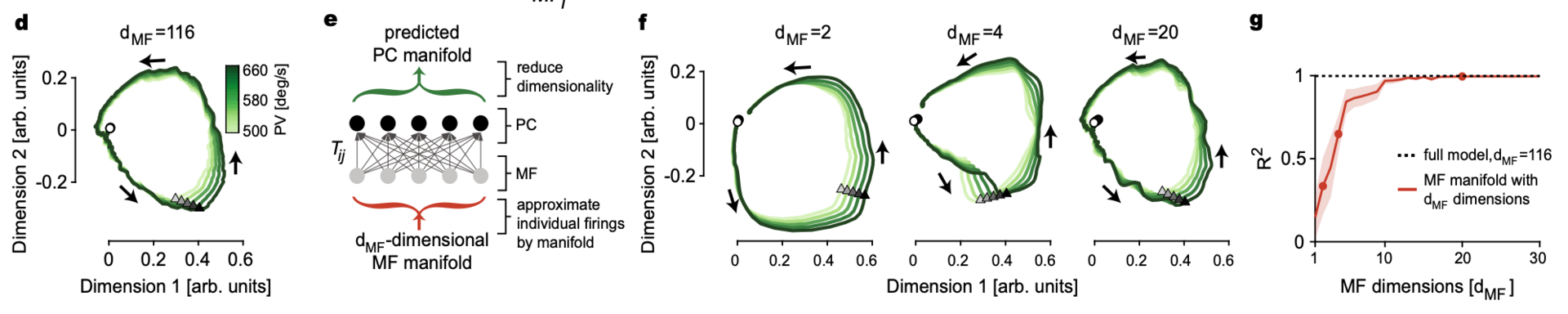
Figure 5: Linear feed-forward network (LFFN) model from MFs to PCs. d LFFN prediction of PC manifolds in Fig. 32d using all 116 dimensions in MF firing. e Schematic diagram of the LFFN model showing steps involved in the prediction of PC manifolds from dMF-dimensional MF manifolds. f Examples of the predicted PC manifold from e when MF manifold dimension is dMF = 2, 4, and 20. g Goodness of fit for the predicted PC manifold to the data versus the input MF manifold dimensions dMF. Dots represent examples in f. Data are mean ± SEM (from Markanday, Hong et al. 2023.
4. Publications
4.1 Journals
- S. Kim, J. Jeon, D. Ganbat, T. Kim, K. Shin, S. Hong and J. Hong: Alteration of Neural Network and Hippocampal Slice Activation through Exosomes Derived from 5XFAD Nasal Lavage Fluid. International Journal of Molecular Sciences 24: 14064.
- J. Myung, S. Hong, C. Schmal, H. Vitel and M.-Y. Wu: Weak synchronization can alter circadian period length: Implications for aging and disease conditions. Frontiers in Neuroscience 17:1242800.
- M.-Y. Kim, M. J. Kim, C. Lee, J. Lee, S.S. Kim, S. Hong, H.T. Kim, J. Seo, K.-J. Yoon and S. Han: Trametinib activates endogenous neurogenesis and recovers neuropathology in a model of Alzheimer’s disease. Experimental & Molecular Medicine 55: 2177–2189.
- Y. Zang and E. De Schutter: Recent data on the cerebellum require new models and theories. Current Opinion in Neurobiology 82:102765.
- M. Kato and E. De Schutter: Models of Purkinje cell dendritic tree selection during early cerebellar development. PLoS Computational Biology 19: e1011320.
- E. De Schutter: Efficient simulation of neural development using shared memory parallelization. Frontiers in Neuroinformatics 17:1212384.
- A. Markanday*, S. Hong*, J. Inoue, E. De Schutter and P. Thier: Multidimensional cerebellar computations for flexible kinematic control of movements. Nature Communications 14: 2548.
4.2 Preprints
- I. Hepburn, J. Lallouette, W. Chen, A.R. Gallimore, S.Y. Nagasawa and E. De Schutter: Hybrid vesicle and reaction-diffusion modeling with STEPS. Preprint at https://www.biorxiv.org/content/10.1101/2023.05.08.539782v1
- A.R. Gallimore, I. Hepburn, S. Rizzoli and E. De Schutter: Dynamic Regulation of Vesicle Pools in a Detailed Spatial Model of the Complete Synaptic Vesicle Cycle. Preprint at https://www.biorxiv.org/content/10.1101/2023.08.03.551909v1
- G. Cirtala and E. De Schutter: Branch-specific clustered parallel fiber input controls dendritic computation in Purkinje cells. Preprint at https://www.biorxiv.org/content/10.1101/2024.01.25.577146v1
4.3 Books and other one-time publications
Nothing to report
4.4 Oral and Poster Presentations
4.4-1 Oral presentations
- S.Hong, Multidimensional cerebellar computations for flexible kinematic control of movement, OIST-RIKEN Brain Symposium 2023, Okinawa, Japan, 21-23 August (2023)
4.4-2. Poster presentations
- G. Cirtala, E. De Schutter, Modeling branch-specific clustered parallel fiber input in a Purkinje cell model with heterogenous ion channel densities, 32nd Annual Computational Neuroscience Meeting, Leipzig, Germany, 15-19 July (2023).
- P. Chou, E. De Schutter, G. Cirtala, Deciphering cerebellar multiplexed coding through the first heterogenous ion channel density branch-specific Purkinje Cell model, Society for Neurosciences (Neuroscience2023), Washington D.C., USA, 11-15 November 2023.
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 OCNC: OIST Computational Neuroscience Course 2023
- Dates: June 19 - July 6
- Venue: OIST Seaside House
- Co-organizers: Kenji Doya (OIST), Tomoki Fukai (OIST), and Bernd Kuhn (OIST)
6.2 Invited talks
- E. De Schutter, Modeling the tripartite synapse at the nanoscale, Department of Neuroscience, Erasmus MC, Netherlands, January 8 (2024).
7. Other
Nothing to report.



