Software
LetItB - Bayesian Estimation tool:Let It Bayesian -
About LetItB
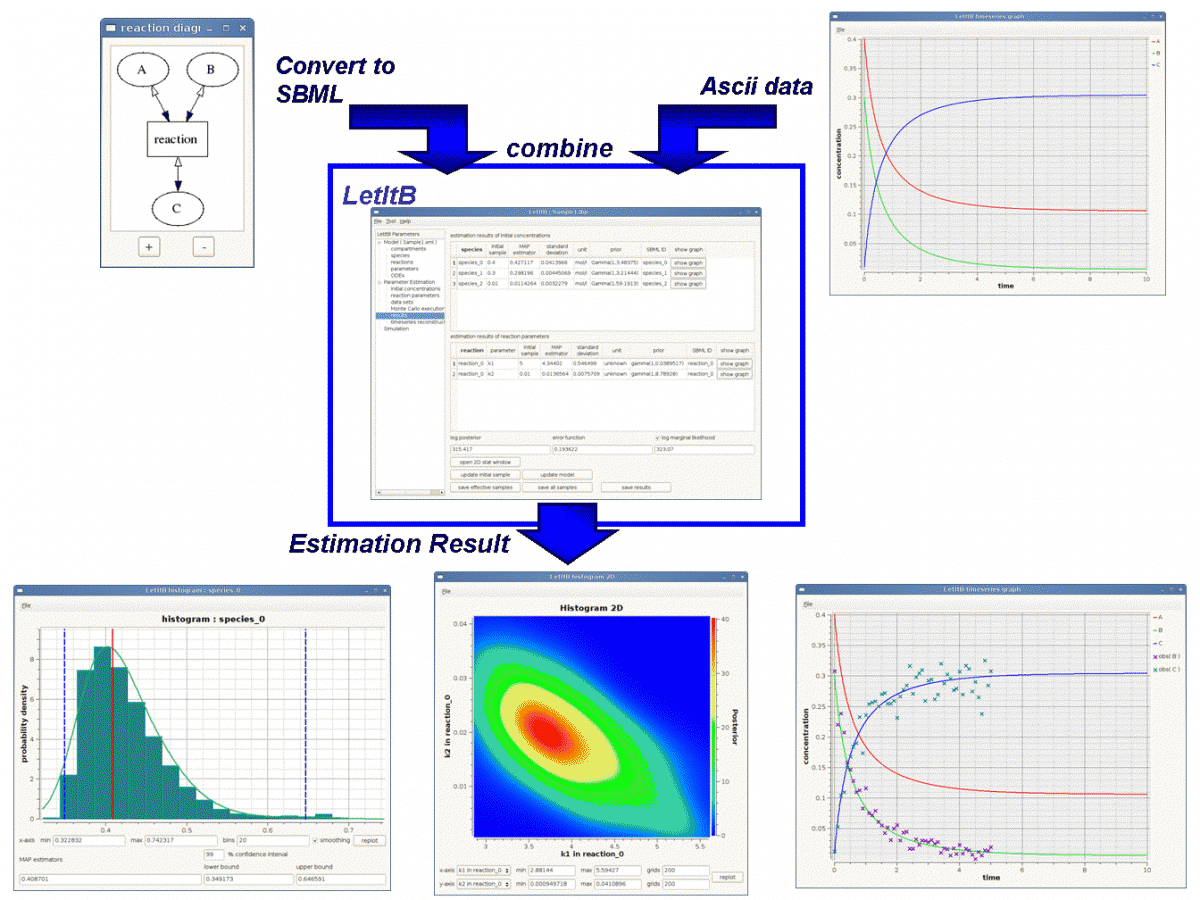
LetItB is developed as a support tool to identify parameters for biochemical reactions. In the current version, LetItB accepts SBML (Systems Biology Markup Language) files as inputs to visually perform the following operations to the SBML models (SBML is the standard format for describing biochemical reaction models in systems biology):
- Create block diagrams to illustrate biochemical reaction networks
- Run simulations
- Identify reaction parameters and estimate their confidence intervals to provide the best recreation for given experimental data (concentration time series of each molecule)
- Assess quantitative model validity from data recreation and model complexity for given experimental data
You can save these results as text files or image files such as jpeg or png via easy-to-use GUI.
LetItB's internal calculation engine is programmed in C language and its Graphic User Interface is developed with the Qt toolkit. Therefore, you can run and recompile LetItB on any platforms including Windows, Mac OS X, and Linux operation systems if you install an appropriate set of libraries in advance.