RNA Aptamer for Use as an RNA-Based Gene Switch That Works in vivo (No. 0222)
|
|
|
<< Back to all technologies |
Summary
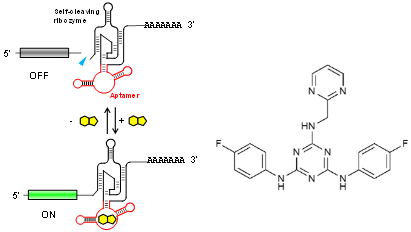
Riboswitches recognize a small molecule through an RNA aptamer and respond by regulating gene expression. Various potential applications for synthetic riboswitches have been proposed and explored in many fields, including gene/cell therapy, regenerative medicine, vaccines, and biologics manufacturing. There have, however, been few reports of small molecules and their cognate aptamers demonstrated to function in mammalian cells, and even those typically require high concentrations of the trigger molecule (~100 μM or higher). A team of OIST researchers led by Prof. Yokobayashi has developed a novel aptamer that binds to a nontoxic compound and works in mammalian cell culture and in vivo in mice at low concentrations. This technology represents a powerful addition to the currently limited toolbox for RNA-based regulation of gene expression.
Applications
- Gene/cell therapy
- Regenerative Medicine
- Vaccines
- Biologics manufacturing
Advantages
- Functional in mammals and mammalian cells
- Regulates gene expression at low conentrations with high ON/OFF ratio
- No exogenous proteins required
- Non-toxic small molecule trigger
Technology
This RNA aptamer was obtained through in vitro selection (SELEX) against ASP7967, an analogue of a well-studied inhibitor of potassium voltage-gated channel subfamily H member 3 (KCNH3). The screening method used ensured that the aptamer is functional in HEK293 cells. Aptazyme-based riboswitches were designed and shown to activate gene expression (>10-fold) in the presence of these compounds at as low as 5 μM in the culture medium. In vivo experimentation demonstrated that an aptazyme-based riboswitch successfully regulated human erythropoietin (hEPO) expression, in response to oral administration of ASP7967 in mice injected with an adeno-associated virus (AAV8) vector. In addition, this riboswitch can be combined with an exon-skipping mechanism to achieve an ON/OFF ratio approaching 300 with an extremely low basal expression level in cultured cells..
Media Coverage and Presentations
CONTACT FOR MORE INFORMATION
![]() Ryohei Yoshida
Ryohei Yoshida
Technology Licensing Section
![]() tls@oist.jp
tls@oist.jp
![]() +81(0)98-966-8937
+81(0)98-966-8937