FY2020 Annual Report
Protein Engineering and Evolution Unit
Assistant Professor Paola Laurino

Abstract
In the past few decades protein engineering allowed to generating artificial enzymes able to catalize unnatural reactions but also to become importnat tools for synthetic biology. Our Unit is particulary interested in generating new enzymes by directed evolution but also to understand how enzymes evolve in Nature.
1. Staff
- Dr. Madhuri Gade, Researcher
- Dr. Bhanu Chouhan, Researcher (till March)
- Dr. Mirco Dindo, Researcher (JSPS Fellow)
- Dr. Saacnicteh Toledo Patino, Researcher
- Dr. Benjamin Clifton, Researcher (JSPS Fellow)
- Dr. Gen-ichiro Uechi, Technician
- Ms. Desirae Martinez, Technician (till August)
- Mr. Stefano Pascarelli, Graduate Student
- Mr. Dan Kozome, Graduate Student
- Mr. Yoshiki Ochiai, Graduate Student
- Mr. Alessandro Bevilacqua, Graduate Student
- Ms. Samira Gmuer, Graduate Student
- Mr. Andrea Testa, strategic JSPS fellow (From February 2020 to August)
- Mr. Aiman Fariz, Intern Student (From February to March)
- Ms. Sachie Matsuoka, Research Unit Administrator(till August)
- Ms. Mika Uehara, Research Unit Administrator
2. Collaborations
2.1 Topic: Discovery of a new metabolite in human cells towards investigation of human methionine adenosyltransferases promiscuities
- Type of collaboration: Joint research
- Researchers:
- Professor Colin Jackson, ANU, Australia
2.2 Topic: Enzymatically Active droplets
- Type of collaboration: Joint research
- Researchers:
- Professor Eric R. Dufresne, ETHZ, Switzerland
2.3 Topic: The functional instability of alanine:glyoxylate aminotransferase
- Type of collaboration: Joint research
- Researchers:
- Professor Barbara Cellini, Perugia University, Italy
- Professor Giorgio Giardina, Rome University, Italy
3. Activities and Findings
3.1 Protein and substrate flexibility contribute to enzymatic specificity in human and bacterial methionine adenosyltransferases
Protein conformational change can facilitate the binding of non-cognate substrates and underlie promiscuous activities. However, the contribution of substrate conformational dynamics to this process is comparatively poorly understood. Here we analyse human (hMAT2A) and Escherichia coli (eMAT) methionine adenosyltransferases that have identical active sites but different substrate specificity. In the promiscuous hMAT2A, non-cognate substrates bind in a stable conformation to allow catalysis. In contrast, non-cognate substrates rarely sample stable productive binding modes in eMAT owing to increased mobility of an active site loop. Different cellular concentrations of substrate likely drove the evolutionary divergence of substrate specificity in these orthologs. The observation of catalytic promiscuity in hMAT2A led to the detection of a new human metabolite, methyl thioguanosine, that is produced at elevated level in a cancer cell line. This work establishes that identical active sites can result in different substrate specificity owing to the combined effects of both enzyme and substrate dynamics.
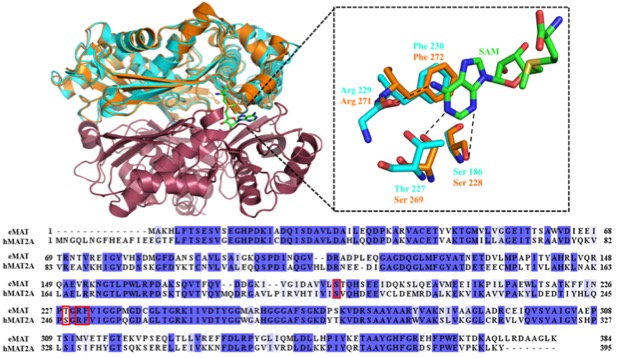
Figure 1: Structural and sequence aligment of MATs.
3.2 Consequences of the divergence of Methionine AdenosylTransferase
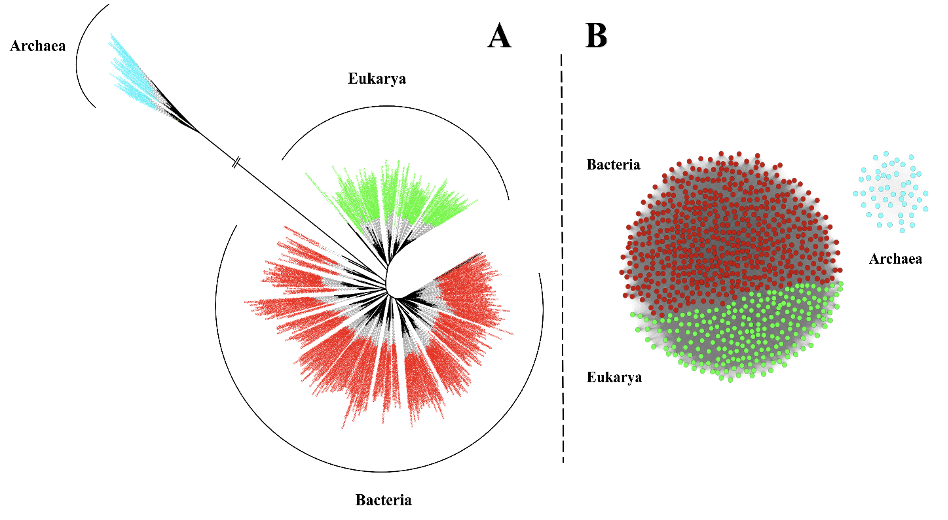
Figure 2: Schematic representation of observed trends in MAT.
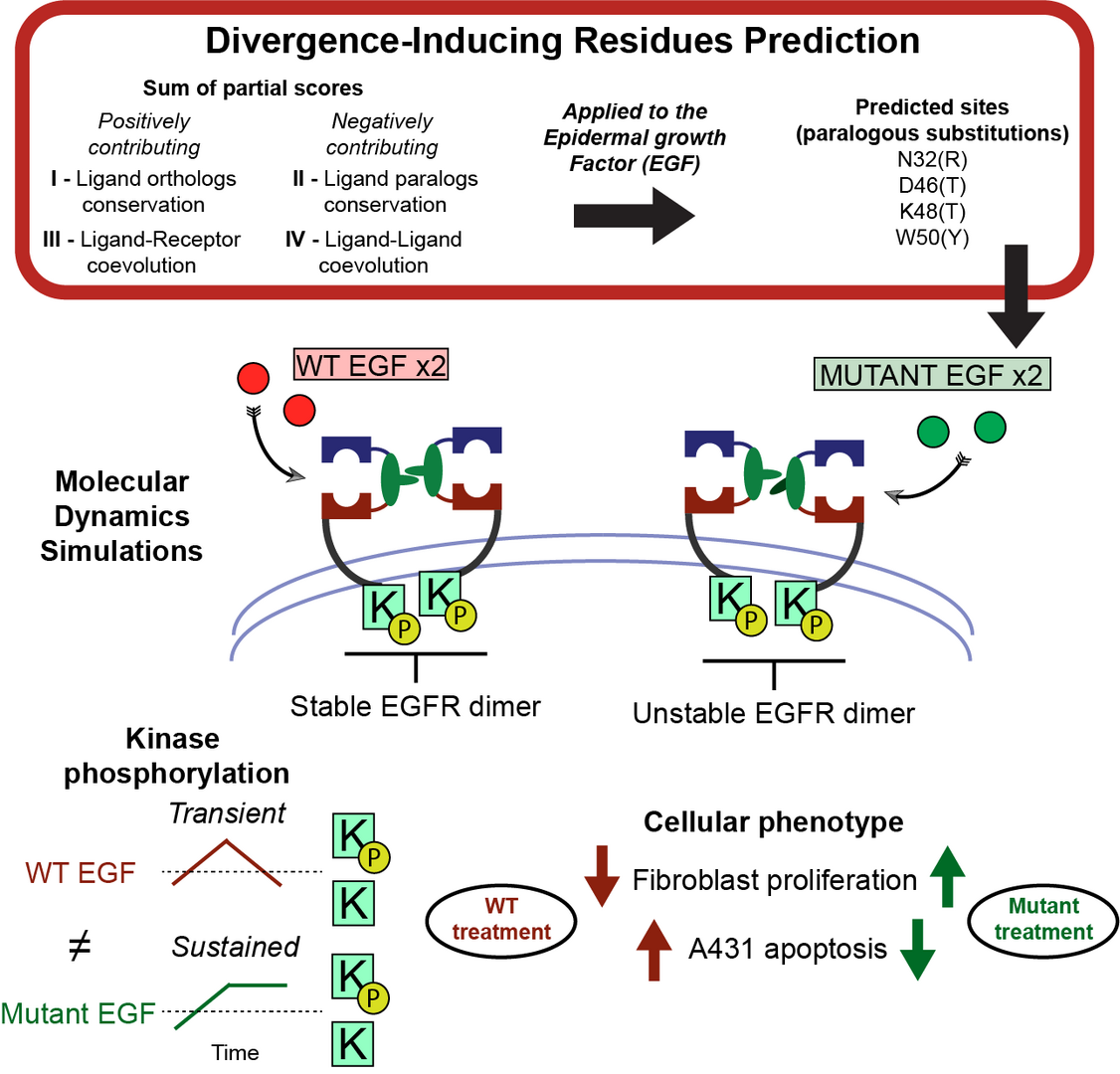
3.3 Single EGF mutants unravel the mechanism for stabilization of Epidermal Growth Factor Receptor (EGFR) system
4. Publications
4.1 Journals
- Danielson, E.*; Dindo, M.; Porkovich, A. J.; Kumar, P.; Wang, Z.; Jain, P.; Mete, T.; Ziadi, Z.; Raghavendra, K.; Laurino, P.; Sowwan, M. Non-Enzymatic and Highly Sensitive Lactose Detection Utilizing Graphene Field-Effect Transistors. Biosensors and Bioelectronics. 2020, 165, 112419 https://doi.org/10.1016/j.bios.2020.112419
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- Benjamin Clifton, “RNA 2020 (25th Annual Meeting of the RNA Society)”, May 26-31, 2020.
- Stefano Pascarelli, "Remote BioExcel Winter School on Biomolecular Simulations", 30 November - 4 December 2020.
- Paola Laurino, “A protein evolution workshop”, 1-3 March 2021 OnLine Volkswagen symposium
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 Seminar: "Harnessing Conformational Dynamics to Engineer New Enzymes"
- Date: July 7, 2020
- Venue: Online
- Speaker: Prof. Lynn Kamerlin, (Uppsala University)
6.2 Seminar: "The reductive glycine pathway - a plug-and-play tool for one-carbon assimilation"
- Date: Aug 18, 2020
- Venue: Online
- Speaker: Dr. Arren Bar-Even, (Max Planck Institute of Molecular Plant Physiology)
6.3 Seminar: "Protein Allostery: Evolution and Correlated Motions"
- Date: Oct 26, 2020
- Venue: Online
- Speaker: Prof. Nozomi Ando, (Cornell University)
6.4 Seminar: "Designing modular proteins"
- Date: Dec 3, 2020
- Venue: Online
- Speaker: Dr. Fabio Parmeggiani, (University of Bristol)
7. Other
1. Dr. Mirco Dindo presented a Science Dialogue demonstration at Kyuyo High School on Jan 27, 2021.
2. Yoshiki introduces OIST as distinct systems from other grad schools in Japan. Published in 実験医学 in Feb 2021.



