Annual Reports FY2015
Neural Computation Unit
Professor Kenji Doya

Abstract
The Neural Computation Unit pursues the dual goals of developing robust and flexible learning algorithms and elucidating the brain’s mechanisms for robust and flexible learning. Our specific focus is on how the brain realizes reinforcement learning, in which an agent, biological or artificial, learns novel behaviors in uncertain environments by exploration and reward feedback. We combine top-down, computational approaches and bottom-up, neurobiological approaches to achieve these goals.
FY2015 was the final year of four of our major externally funded research projects and we made remarkable progress in toward their conclusion.
In the Kakenhi project on Prediction and Decision Making, we showed that the neural circuit parietal cortex realizes dynamic Bayesian inference through analysis of two-photon neural imaging data collected in collaboration with Kuhn Unit at OIST (3.2.1). We also started to introduce new neural imaging technology using a miniature endoscope microscope (3.2.2, 3.2.3).
In the Strategic Research Program for Brain Sciences (SRPBS), Theme F, we developed machine learning methods for subtype identification and diagnosis of depression from resting-state fMRI and other multidimensional data in collaboration with Prof. Yamawaki's Lab at Hiroshima University (3.1.1). We also showed that facilitatory effect of optogenetic activation of serotonin neurons on waiting for delayed rewards is dependent on the probability of reward delivery (3.2.4).
In SRPBS, Theme G, in collaboration with Prof. Kaibuchi's lab at Nagoya University, we opened a database KANPHOS regarding phosphoproteomics of dopamine-related signaling pathways (3.1.2). We also created a signaling cascade model of dopamine D2 receptor expressing striatal neurons including adenosine receptors (3.1.3).
In the Supercomputational Life Science Program, we developed a large-scale spiking neuron model of the basal ganglia-thalamo-cortical loops and, in collaboration with Profs. Takagi and Nakamura's labs at University of Tokyo, realized an integrated simulation with the spinal cord and the musculoskeletal system to reproduce the symptoms of Parkinson's disease (3.1.4).
We also made new developments in the theory of inverse reinforcement learning (3.3.1) and the framework of reinforcement learning using Restricted Boltzmann Machines (3.3.2).
1. Staff
Dynamical Systems Group
- Junichiro Yoshimoto, Group Leader
- Jun Igarashi, Staff Scientist
- Jan Moren, Staff Scientist
- Naoto Yukinawa, Staff Scientist
- Ildefons Magrans de Abril, Staff Scientist
- Carlos Enrique Gutierrez, Postdoctoral Scholar
- Hiromichi Tsukada, Postdoctoral Scholar
- Jessica Verena Schulze, OIST Student
- Kosuke Yoshida, Special Research Student
Systems Neurobiology Group
- Makoto Ito, Group Leader
- Akihiro Funamizu, Postdoctoral Scholar
- Kazumi Kasahara, JSPS Research Fellow
- Katsuhiko Miyazaki, Staff Scientist
- Kayoko Miyazaki, Staff Scientist
- Yu Shimizu, Staff Scientist
- Hiroaki Hamada, OIST Student
- Tomohiko Yoshizawa, Special Research Student
Adaptive Systems Group
- Eiji Uchibe, Group Leader
- Stefan Elfwing, Researcher
- Qiong Huang, OIST Student
- Shoko Igarashi, OIST Student
- Farzana Rahman, OIST Student
- Chris Reinke, OIST Student
- Jiexin Wang, Special Research Student
- Matti Kruger, OIST Student
- Paavo Parmas, OIST Student
- Tadashi Kozuno, OIST Student
Administrative Assistant / Secretary
- Emiko Asato
- Kikuko Matsuo
2. Collaborations
2.1 Development of reinforcement learning technology for large scale systems and its application to ICT systems
- Type of collaboration: Joint research
- Partner organization: Fujitsu Laboratories Ltd.
- Researchers:
- Seishi Okamoto, Fujitsu Laboratories Ltd.
- Naoki Sashida, Fujitsu Laboratories Ltd.
- Tomotake Sasaki, Fujitsu Laboratories Ltd.
3. Activities and Findings
3.1 Dynamical Systems Group
3.1.1 Subtype identification and diagnosis of depression from multidimensional data using machine learning [Shimizu, Tokuda, Yoshida, Yoshimoto]
Diagnosis of depression is currently based on long interviews. In the search for an objective and more efficient method to diagnose this complex disease we analyze multidimensional data, including structural magnetic resonance imaging (MRI) data, functional MRI data involving four different tasks, resting state data, as well as blood markers, genetic polymorphism and behavioral tests, obtained from depression patients and healthy controls using supervised and unsupervised machine learning approaches.
In the supervised learning approach, we have shown that suitable application of L1 sparse logistic regression to be an effective tool for distinction of patients and the relevant characteristics of the investigated data (Shimizu et al., 2015). Application of this method to further data revealed, that depression patients can be characterized with 72% accuracy by means of the functional connection between six brain areas involved in the default mode network (Table 1), as well as 76% accuracy by means of the methylation rate at specific depression related genetic sites (Table 2).
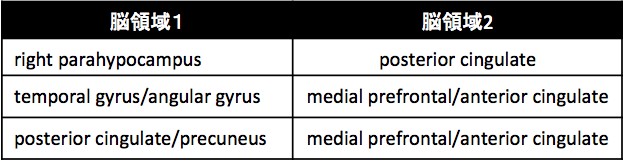
Table 1: Brain areas, for which functional connection showed to be stronger in depression subjects than healthy controls.
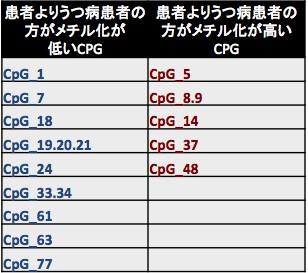
Table 2: CpGs, for which the methylation rate was distinctive for depression patients.
In the unsupervised learning, we developed a multiple co-clustering method that is applicable to concatenated data with different types of features such as numerical and categorical. Unlike a conventional clustering method, this novel method can simultaneously perform feature selection and cluster analysis. Hence, this enables us to identify a cluster structure characterized by a small number of features, which would be otherwise hidden in the conventional clustering approach. Next, we applied this method to our dataset, which consisted of functional MRI, clinical questionnaires, and blood markers. As a result of analysis, a specific cluster solution was identified as relevant for differences between healthy and depressive subjects. Further, in this clustering solution, two clusters were found relevant for healthy subjects while three were for depressive ones. Furthermore, these three depressive clusters were characterized by specific features: treatment effect of anti-depressive drug; stressful experiences in childhood; functional connectivity in default mode networks in the brain (Fig.3.1. 1). These results imply possible diagnosis of treatment effect, based on stressful experiences in childhood and function connectivity in default mode network, providing a novel approach to diagnosis of subtypes of depression in practice.
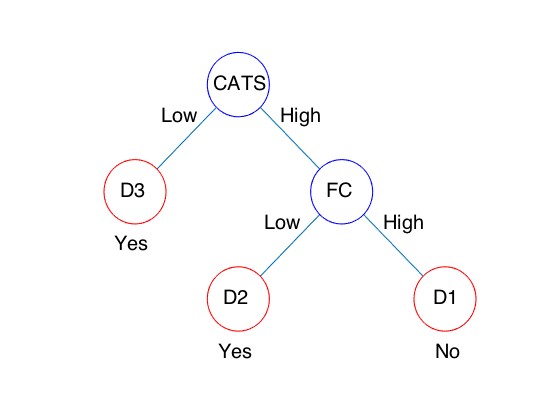
Fig.3.1.1 Classifier of depressive subjects based on CATS(stressful experiences in childhood) and FC(functional connectivity). When a subject is classified into D1, anti-depressive drug (SSRI) may not be effective. On the other hand, when a subject is classified into either D2 or D3, the drug is effective.
3.1.2. A neuroinformatics platform for protein phosphorylation with quality control [Yoshimoto]
Protein phosphorylation is involved in pathways of a wide variety of physiological processes in the nervous system. Nonetheless, little is known about which sites are phosphorylated by a specific regulator (kinase) and which extracellular stimuli activate (or inhibit) the protein phosphorylation via intracellular signaling cascades. To uncover the basic issue, our collaborators, Prof. Kozo Kaibuchi and colleagues at Nagoya University Graduate School of Medicine, developed a new methodology for screening the target phosphorylated sites of a given kinase (or extracellular stimulus) using the mass spectrometry, and succeeded in identifying hundreds of phosphorylated sites of representative kinases (e.g. PKA, MAPK, Rho-Kinase, etc.). To summarize the data systematically and extract biologically significant information, we have been developing a database system named KANPHOS (Kinase-Associated Neural Phospho-Signaling) for these years.
The database system and its web portal were built based on XooNIps, and have been released since March 2016 (Fig. 3.1.2.1). All data are controlled for quality via review and curation by specialists. The web portal supports three modes of search: 1) Search for substrates phosphorylated by a specific kinase; 2) Search for kinases phosphorylating a specific protein; and 3) Search for kinases and their target substrates by a specific signaling pathway. Each protein (kinase/substrate) item is linked with 15 external databases, enabling us to easily predict unknown functions of the protein phosphorylation. Also, they provide a function to show a list of pathways in which the set of substrates phosphorylated by a specific condition is overrepresented more than expected, via communication with Reactome (http://www.reactome.org). Prediction based on this function and pharmacological experiments uncovered that the Rap1 signaling in D1-type receptor expressing medium-sized spiny neurons (D1R-MSNs) is activated by dopaminergic stimulus, thereby the excitability of D1R-MSNs is enhanced. Those results were published in (Nagai, et al., 2016, See also Fig. 3.1.2.2).
[Reference]
Nagai, T., …, Takano, T., Yoshimoto, J., …, Kaibuchi, K. (2016). Phosphoproteomics of the Dopamine Pathway Enables Discovery of Rap1 Activation as a Reward Signal In Vivo. Neuron, 89(3), 550–565.

Fig. 3.1.2.1: Appearance of KANPHOS web potal site (https://kanphos.neuroinf.jp/)
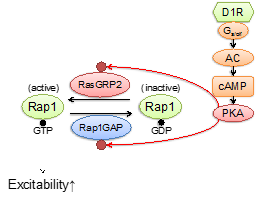
Fig. 3.1.2.2: Pathways in D1-type receptor expressing medium-sized spiny neurons uncovered based on the KANPHOS database.
3.1.3 Development of a multiscale simulation model of the cortico-basal ganglia circuit [Yukinawa, Yoshimoto]
The cortico-striatal pathways coordinately play a fundamental role for the emotional response and action selection of animals. Several key regulatory molecules including dopamine and adenosine are known to modulate the excitability by controlling the intracellular signaling pathways of the striatal medium spiny neurons (MSNs). However, the detailed quantitative actions and the effect to the cognitive/motor functions remain largely unresolved. To predict the pharmacokinetic capabilities of the molecules from the cellular level to the whole circuit level, we had developed a multiscale simulation model of the cortico-basal ganglia system for last two fiscal years in the Brain/MINDS project (AMED, Japan).
In this final fiscal year, we refined the model by introducing realistic morphological structures of the dopamine D2 receptor expressing MSNs (D2R-MSNs). The updated model includes a detailed multicompartment neuron model of the D2R-MSN which has region-dependent distribution of the ion channels, reconstructed from a stack of fluorescence microscopic images of a D2R-MSN (Fig. 3.1.3.1 (A)). We also updated the signal transduction model of the D2R-MSN. The model assumes that the cellular excitability is modulated by the common PKA-RasGRP2/Rap1GAP-RAP pathway confirmed in the D1R-MSNs (Fig. 3.1.3.1 (B)). By using the model, we showed that administration of D2R antagonist activates PKA and results in enhanced phosphorylation of Rasgrp2, which is consistent with an experimental result. We further predicted that the concentration of extracellular adenosine dynamically controls the phosphorylation level (Fig. 3.1.3.1 (C)).
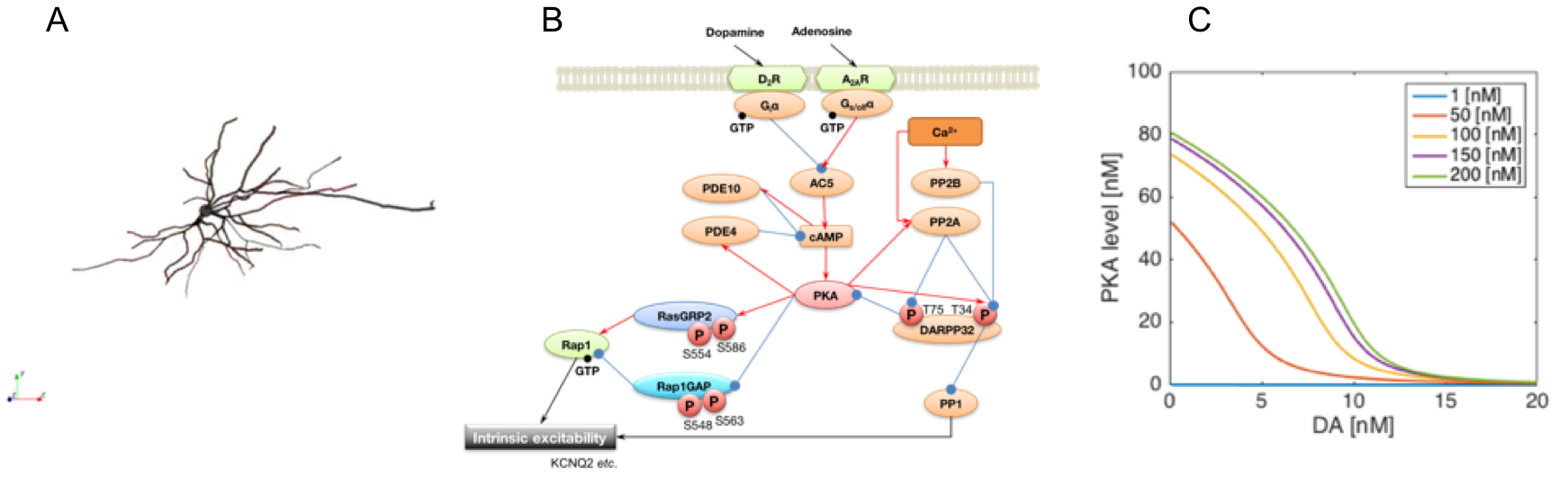
Fig. 3.1.3.1: A. Reconstructed 3D morphology of a D2R-MSN. B. The updated signal transduction model of excitability regulation in the D2R-MSN. C. Simulated dose-response curves of active PKA for dopamine inputs.
By incorporating these model elements, we constructed an integrated simulation model of the cortico-basal ganglia circuit which combines the different biological layers including the signal transduction, cells, neuronal circuits, and behavior (Fig. 3.1.3.2 (A)). In the model, activities of the D1R-/D2R-MSNs in the nucleus accumbens (NAc) are integrated at the substantia nigra pars reticulata (SNR) through the direct/indirect pathways and thus the modulation of each neuronal population will have major influences on the behavioral frequency. We used the model to predict effect of the pharmacological action of biologically plausible concentration levels of tonic extracellular dopamine and adenosine on behavior. Focusing on relative difference between active PKA levels of D1R-MSNs and D2R-MSNs as criterion for behavioral frequency, the results show that adenosine substantially modulates the sensitivity of behavior to dopamine (Fig. 3.1.3.2 (B)) and that adjusting the adenosine level under a wide range of dopamine concentration conditions can normalize the balance between direct and indirect pathways by modulating excitability of the NAc to physiological levels (Fig. 3.1.3.2 (C)). These results may explain the quantitative basis for the effectiveness of adenosine A2A receptor antagonists which improve some movement symptoms in Parkinson's disease, one of dopamine related diseases.
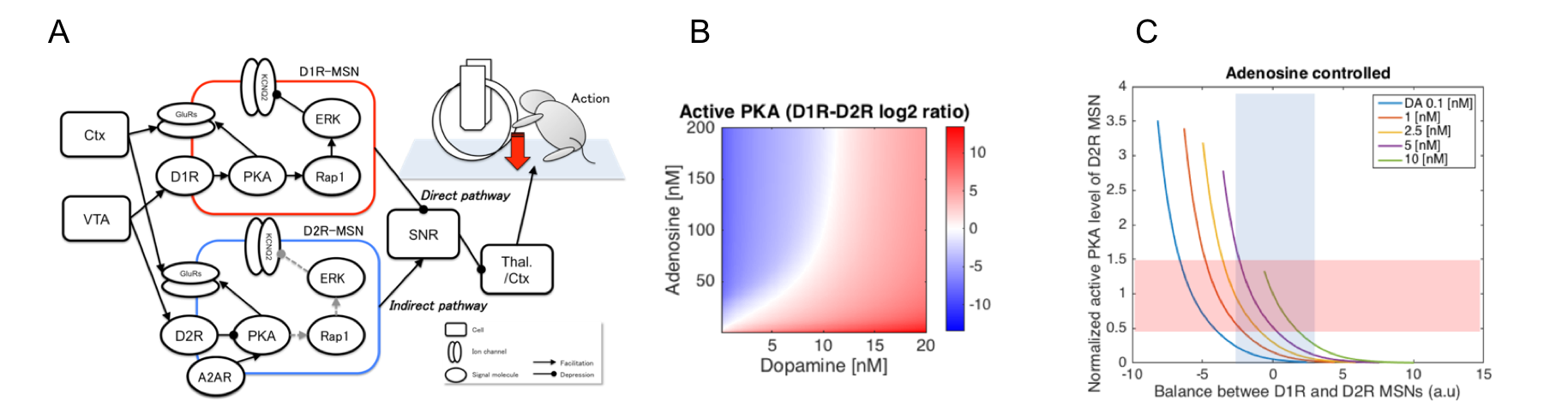
Fig. 3.1.3.2: A. Integrated simulation model of the cortico-basal ganglia circuit. B and C. Simulated activation balance between D1R and D2R MSNs in different extracellular signal configurations at the biological ranges. Modulation of extracellular adenosine improves wide range of low dopaminergic states with respect to balance and signal level (> 1 nM DA).
3.1.4 Large-scale spiking neuron models of the basal ganglia-thalamo-cortical circuit [Moren, Igarashi, Shouno, Yoshimoto]
The basal ganglia are the locus of Parkinson's disease and the basal ganglia-thalamo-cortical circuit is known to play a critical role in the development of tremor and other motor-related symptoms. However, the precise mechanism for the loss of dopaminergic projection to the basal ganglia circuit to cause Parkinsonian tremor is still unknown. To understanding the dynamic circuit mechanisms of Parkinsonian pathology, we constructed an integrated model of the basal ganglia-thalamo-cortical circuit by FY2014.
In the final year of the project, we worked on integrating the brain model with the spinal cord and musculoskeletal system model constructed by our collaborators at Takagi and Nakamura labs in University of Tokyo. By using MUSIC (multi-simulation coordinator), we successfully ran the integrated simulation on the K supercomputer.
3.2 Systems Neurobiology Group
3.2.1 Neural substrate of dynamic Bayesian inference in the posterior parietal cortex [Akihiro Funamizu, collaboration with Professor Kuhn]
Our brain often receives limited sensory information to understand the state of the outside world. Therefore the ability to estimate the current state through mental simulation is essential. In a stochastic dynamic environment, this state estimation is realized by dynamic Bayesian inference, such as Kalman filtering. To investigate the neural substrate of dynamic Bayesian inference, we use two-photon imaging in awake behaving mice which enables us to simultaneously image multiple identified neurons and their calcium activities while the mouse conducts an auditory virtual navigation task.
A mouse is head restrained and maneuvers a spherical treadmill. 12 speakers around the treadmill provide an auditory virtual environment. The direction and amplitude of sound pulses emulate the location of the sound source, which is moved according to the mouse’s locomotion on the treadmill. When the mouse reaches the sound source (goal) and licks a spout, it gets a water reward. The task consists of two conditions: continuous condition in which the guiding sound is presented continuously and intermittent condition in which the sound is presented intermittently.
In the task, mice increase licking as they approached the goal both with and without cue sounds. The anticipatory licking is disturbed selectively during sound suspension by inactivating posterior parietal cortex (PPC) with the GABA-A receptor agonist muscimol. These results suggest that mice estimate the goal distance based on their own actions and that PPC is involved in the estimation.
Calcium is an important second messenger in neurons and an increased calcium concentration is correlated with neuronal activity. For this reason we use the genetically encoded calcium indicator GCaMP6f in PPC and adjacent posteromedial cortex (PM) of mice to detect neuronal activities with two-photon microscopy. We record the activities of 200 – 500 neurons simultaneously in each of layers 2, 3 and 5 for one hour continuously (Figure 3.2.3). By decoding the population activity using machine learning techniques, we find that some neurons represented goal distances during sound suspension (prediction), more robustly in PPC than in PM and in layers 3, 5 more robustly than in layer 2. Uncertainty of prediction decreased with observation of cue sounds (updating). These results suggest that cortical microcircuits implement dynamic Bayesian inference for state estimation and that PPC takes a role in the estimation of target location using an action-dependent state transition model.
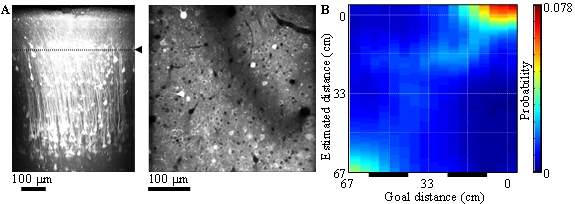
Figure 3.2.1: A. In-vivo imaging of neurons with two-photon microscopy. Reconstruction of cortical neurons in posterior parietal cortex (left). Dotted line indicates the imaging plane of layer 2 (right).
B. Distance estimation with a probabilistic decoder in layer-2 PPC neurons during intermittent condition. Horizontal and vertical axes show the actual and estimated distance to sound source, respectively. Bars below show sound zones. The estimations were successful even during no-sound zones.
3.2.2 CA1 pyramidal neurons encode not only spatial information but also moving speed of an animal: information extraction by an unsupervised-learning method [Makoto Ito]
With the recent progress in neural recording technology, especially in calcium imaging, the number of cells that can be recoded simultaneously has been steadily increasing. Accordingly, how to extract the information coded in large population activities has become an important issue.
For instance, it is well known that the hippocampus represents where an animal is in its environment. Using a data set of the trajectories of a moving animal and the hippocampal population activities recoded at the same time, previous studies have designed decoders that estimated a corresponding place of the animal for a given population activity. This decoding method is useful to quantify the information that the neural population codes, but it is impossible to find out unknown information that researchers don’t think of.
In this study, we showed that a nonlinear unsupervised-learning method, t-distributed stochastic neighbor embedding (t-SNE), could extract not only spatial information but also unexpected information correlating with the moving speed, only from neuronal data without using trajectory data.
We recorded activities of 338 CA1 pyramidal neurons expressing calcium indicator (GCaMP6f) with a miniature fluorescence microscope (inscopix), while a rat was exploring in an open space (45 cm x 45 cm). We applied t-SNE to the population activities, which are regarded as the points in a high-dimensional space spanned by each neuron’s activity, to extract a lower- (three-) dimensional structure where the probabilistic distances between each point are preserved as much as possible. Then, we added a new coordinate system for the structure by a principal component analysis.
We found that 1st and 2nd components represented the position of the rat in the environment. After the adequate transformation (mirror, rotate, and scale), the mean-squared error from the x-y trajectories was significantly lower than the chance level (7.2 cm < 22.5 cm). Furthermore, we found that the remaining 3rd component significantly correlated with the moving speed of the rat. When the 3rd component was a lower value, the tendency of staying at the same position was stronger.
These results demonstrated that an unsupervised-learning method can be a strong tool to extract unknown information from the population activity. By this method, we newly found that the moving speed was also represented in the hippocampus as well as spatial information.
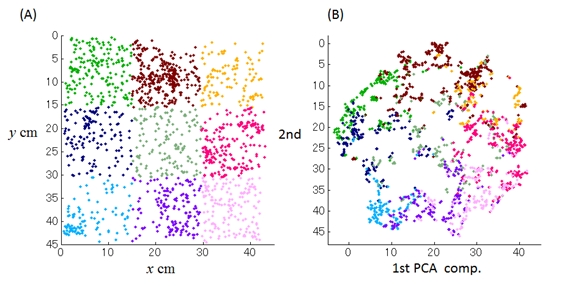
Figure 3.2.2: (A) Trajectory of a rat in a 45x45cm box. (B) Applying mirror, rotate, and scale transformation, we found that 1st and 2nd components of population activity of 338 CA1 neurons well match up nicely with the rat’s position.
3.2.3 The striatal striosome neurons encode the value of sensory stimulus [Tomohiro Yoshizawa]
The striatum consists of the striosome (patch) and matrix compartments that make mosaic structure in the striatum. Anatomically, the striosome receives the projection from the limbic cortex, whereas the matrix receives projections from the sensorimotor and associative cortex. The D1-type medium spiny neurons in the striosome compartment directly projects to the dopaminergic neurons of the substantia nigra pars compacta (SNc). Although the striatum is involved in value-based learning, the specific roles of these compartments has not been clear because the mosaic structure makes it difficult to identify the compartments during electrode recording. We previously hypothesized that the striosome compartment represents the state value, whereas the matrix represents the action value in reinforcement learning (Doya, 2000). In this study, to test whether the striosome neurons represent value information or not, we conducted endoscopic deep-brain in vivo calcium imaging (nVistaHD, Inscopix) of transgenic mice with selective GCaMP6s expression in the striosome neurons (Gerfen et al., 2013).
Mice were classically conditioned with four odor cues predicting reward, aversive stimuli, or nothing. Each trial began with a conditional stimulus (CS; odor), followed by a delay period and an unconditional stimulus (water/air puff/nothing). Within the first 7 days, reward-predictive behavior was formed for the odor stimuli associated with rewards. We recorded the activity of the same striosome neurons over 15 days and compared the activities in early learning stage with late learning stage. As the results, we found two types of striosomal value-coding neuron depending on the task learning stage. Type I value-coding neurons showed activities that modulated by the upcoming reward amounts when reward predictive behavior appeared (= early learning stage). Type II value-coding neuron encoded value information when mice mastered the odor-reward association (= late learning stage). And the proportion of the proficiency type in the striosome was larger than in the all striatum.
The existence of stirosome neurons coding value information was revealed for the first time using selective optical recording. The result suggests that one of the roles of striosome neurons is to associate sensory states with predicted reward, in consistence with our hypothesis about value coding in the striosome compartment.
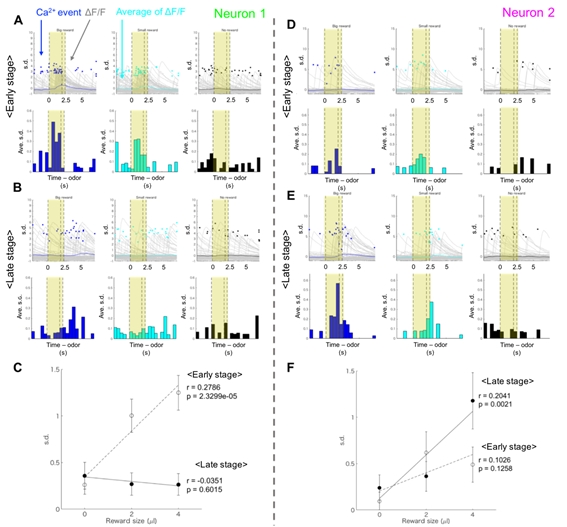
Figure 3.2.3: (A, B) An example of striatal neural activity modulated by the upcoming reward amount in the early learning stage (Type I value-coding neuron). Yellow areas indicate the CS and delay period. (C) Averaged activities of Neuron 1 in the CS-delay period. The significant correlation was observed between the reward size and the neural activity in the early learning stage. (D, E) An example of striosomal neural activity modulated by the upcoming reward amount in the late learning stage (Type II value-coding neuron). (F) Averaged activities of Neuron 2 in the CS-delay period. The significant correlation was observed between the reward size and the neural activity in the late learning stage.
3.2.1 The role of serotonin in the regulation of patience [Katsuhiko Miyazaki, Kayoko Miyazaki]
While serotonin is well known to be involved in a variety of psychiatric disorders including depression, schizophrenia, autism, and impulsivity, its role in the normal brain is far from clear despite abundant pharmacology and genetic studies. From the viewpoint of reinforcement learning, we earlier proposed that an important role of serotonin is to regulate the temporal discounting parameter that controls how far future outcome an animal should take into account in making a decision (Doya, Neural Netw, 2002).
In order to clarify the role of serotonin in natural behaviors, we performed neural recording, microdialysis measurement and optogenetic manipulation of serotonin neural activity from the dorsal raphe nucleus (DRN), the major source of serotonergic projection to the cortex and the basal ganglia.
We introduced transgenic mice that expressed the channelrhodopsin-2 variant ChR2(C128S) in the serotonin neurons. We found that serotonin neuron stimulation prolonged the time animals spent for waiting in reward omission trials (Figure 3.2.4). This effect was observed specifically when the animal was engaged in deciding whether to keep waiting and not due to motor inhibition. We also found that serotonin effect on promoting waiting time occurred only when reward probability was high. These results established a causal relationship between serotonin neural activation and patient waiting for future rewards.
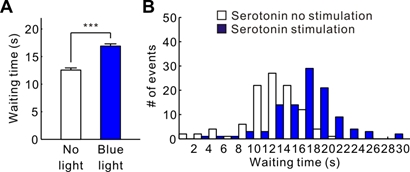
Figure 3.2.4: (A) The mean waiting time during the omission trials in serotonin no stimulation or in serotonin. (B) The distribution of the waiting times during the omission trials in (A). ***p < 0.001.
3.2.5 Biological elucidation of neural plasticity by motor training [Kazumi Kasahara, Collaboration with Dr. Hanakawa, IBIC, NCNP]
The potential of brain-machine interfaces (BMIs) to replace lost neuronal function in the form of neuroprostheses has been widely studied. Moreover, BMIs have gained recent attention as a possible means to induce beneficial neuronal plastic changes via neurofeedback training. However, although BMI performance considerably varies among subjects, the factors affecting BMI performance are poorly understood. Therefore, we investigated the relationship between performance of an electroencephalographic (EEG) mu rhythm-based BMI and brain structure. We found that BMI performance correlates of brain volumes in dorsal premotor cortex (PMd), supplementary motor area (SMA), and supplementary somatosensory area (SSA) (Kasahara, et al., Neuroimage 2015). This finding indicates that control performance for the present EEG-BMI was higher in subjects with greater these volumes. Specially, the PMd is known to be involved in motor imagery, and we speculate that subjects with greater PMd volume were more adept at imagining hand movement. This result may help to develop “Taylormade-BMI” suiting each subject and to induce beneficial neuronal plastic changes. Therefore, this finding may help the development of BMI. This research was collaborated with Dr. Takashi Hanakawa, director of department of Integrative Brain Image center (IBIC), National Center of Neurology and Psychiatry (NCNP).
In addition, recent studies have demonstrated that longitudinal motor training induced to beneficial changes of brain volume and fiber structure using Magnetic resonance imaging (MRI). However, its biological mechanism behind the plasticity remains unknown. The aim of our next step is, using both MRI and histological analyses, to elucidate the biological evidence of neuronal plastic changes induced by motor training. The elucidation would contribute to not only advances in science but also developments of novel therapies for neurological disorder like BMI and neurofeedback training. Therefore, we are now training mice to push button sequentially to induce beneficial plasticity in motor area. This experiment is supported by Ms. Jiafu Zeng who is a technician.

Figure 3.2.5: BMI system and gray matter volumes (red clusters; PMd, SMA, and SSA) which correlated of BMI performance. Based on their electro-encephalogram (EEG), patients or healthy volunteers are able to move their computer cursor, wheel chair, care robot, and so on.
3.3 Adaptive Systems Group
3.3.1 Deep inverse reinforcement learning by logistic regression [Uchibe]
Inverse Reinforcement Learning (IRL), which is a method of estimating a reward function that can explain a given agent's behavior, provides a computational scheme to implement imitation learning. We proposed the IRL that can estimate the reward and the value functions at the same time (Uchibe and Doya, 2014), which have been already reported in the annual report in FY2014. This study extends our previous method by introducing deep learning frameworks to identify the nonlinear representation of reward and value functions. Since our IRL algorithm is formulated as a binary logistic regression problem, the application of deep learning frameworks is straightforward, and the network structure of a binary classifiers is derived from the simplified Bellman equation under linearly solvable Markov decision process (Todorov, 2007; 2009). Figure 3.3.1 shows the network architecture, that consists of three networks: density ratio, reward, and value function. The goal of the classifier is to classify whether the data are sampled from the optimal transition probability or baseline probability.
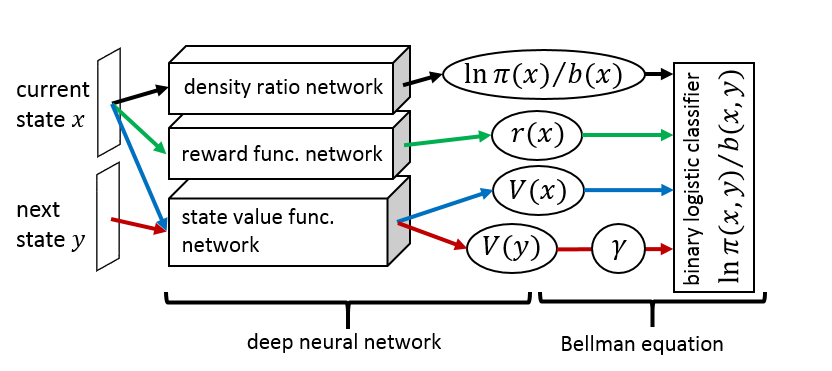
Figure 3.3.1: Proposed network architecture for inverse reinforcement learning that consists of three networks: density ratio, reward, and value function. Then the Bellman equation is computed from the outputs of the three networks.
We selected the game of Reversi (a.k.a. Othello) to evaluate the proposed method and the previous method proposed by Wulfmeier et al., (2016). To collect the dataset from the optimal transition probability, we prepared three stationary policies (RANDOM, HEUR, and COEV) used in previous studies, and every policy repeatedly plays against every other. Then the state transitions are retrieved from the game trajectories of the winners. On the other hand, the dataset of the baseline probability is constructed by retrieving the state transitions from the trajectories in which two RANDOM policies play the game. After optimizing the classifier, the reward function can be retrieved from the parameters of the classifier.
Since our method estimates the value function at the same time, it is used as the potential function for reward shaping to accelerate speed of learning. The optimal policy trained with the shaping reward outperformed three stationary policies and optimal policies trained by Wulfmeier's method. This study will be appeared in ICONIP 2016.
3.3.2 Expected Energy-based Reinforcement Learning [Elfwing]
We have reported the Expected Energy-based Restricted Boltzmann Machine (EE-RBM) for classification in the FY2014 annual report. In this study, we apply the EE-RBM to approximate the state-action value function of the reinforcement learning algorithm.
We selected Stochastic SZ-Tetris (Burgiel, 1997) as a benchmark task to validate our EERL. We consider three learning settings: (a) TD(l) with state features, (b) SARSA(l) with state features, and (c) TD(l) with board states. Note that (a) and (c) are model-based approaches, and (b) is model-free one, respectively. The state features are used in previous study (Bertsekas and Ioffe, 1996) while the board states represent the state of the board as a kind of image like deep learning literatures. EERL was compared with Free Energy based Reinforcement Learning (FERL) and Neural Network based RL (NNRL). As shown in Figure 3.3.2, EERL worked well for all the three learning settings. Interestingly, NNRL could not find the policy for the model-free setting (b) and the learning with board states (c).
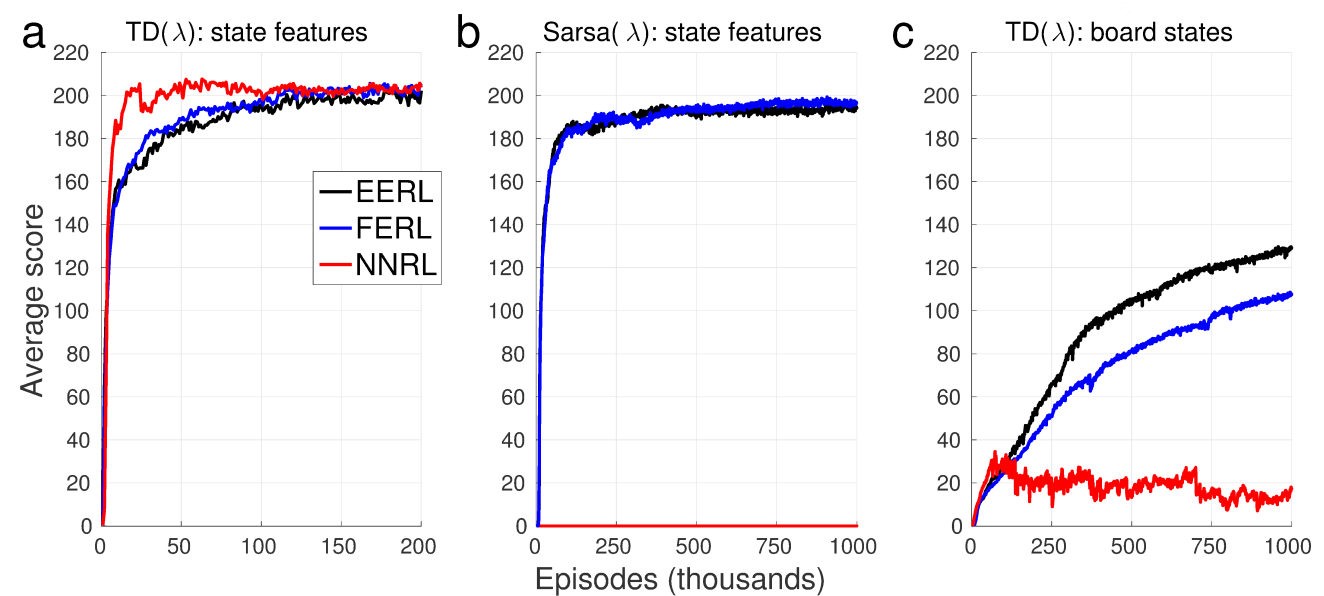
Figure 3.3.2: The average score computed over every 1000 episodes and 5 simulation runs in the three learning settings for SZ-Tetris: (a) TD(l) with state features, (b) SARSA(l) with state features, (c) TD(l) with board states.
3.3.3 Analysis of the elite parameter of EM-based policy hyper parameter exploration [Wang]
In FY2014, we proposed the EM-based Policy Hyper parameter Exploration (EPHE) method for finding an optimal deterministic policy, that is a combination of Policy Gradient Parameter Exploration (PGPE) and Reward Weighted Regression. The advantage is that our method does not need to tune the learning rate, which significantly reduce the number of preliminary experiments to find appropriate meta-parameters of learning algorithms..
Our method is interpreted as evolutionary computation, and we only take the parameters from the best K samples in N trajectories for updating to obtain a good sampling performance. This heuristic works well in practice, but we need to tune this elite parameter K manually. Therefore, we investigate how the elite parameter affects the learning performance in the inverted pendulum swinging, cart-pole balancing, and our smartphone robot standing-up and balancing tasks in computer simulation.
Figure 3.3.3 shows the sensitivity of K to N in the three tasks. Agents with smaller setting of K learned relatively faster but reached no better performance in the end. There was no significant difference between different settings of K, but agents without the selection mechanism achieved much worse behaviors.
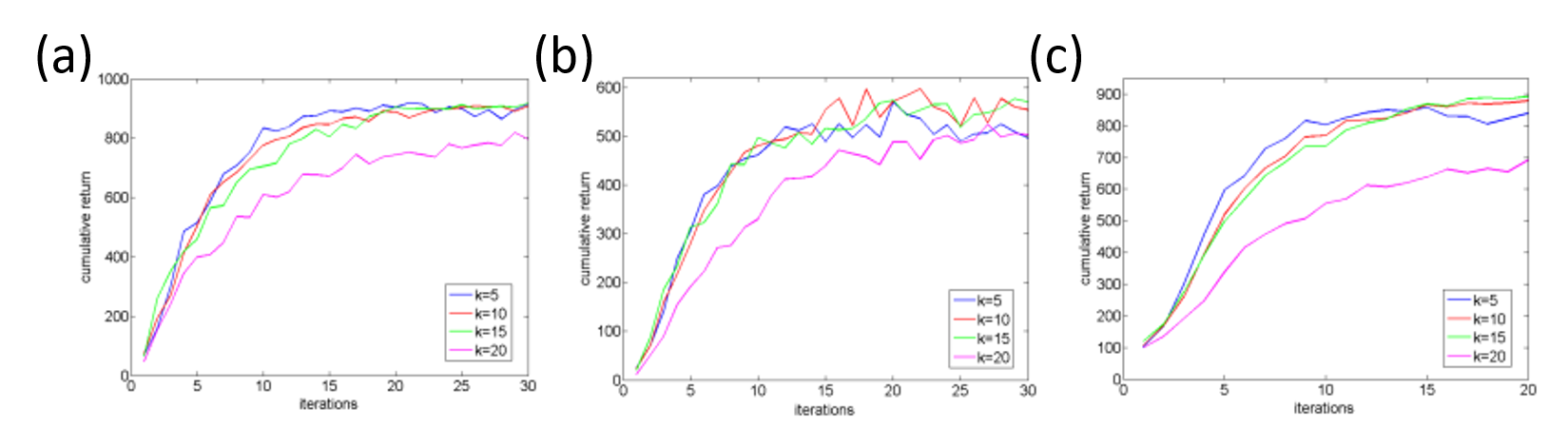
Figure 3.3.3: Sensitivity of the elite parameter K to the number of samples N: (a) Swing-up the inverted pendulum task, (b) Cart-pole balancing task. (c) Smartphone robot standing-up and balancing task.
3.3.4 Gamma-Ensemble for maximizing average reward [Reinke]
Based on the neuroscientific findings, Kurth-Nelson & Redish (2009) proposed a dependent γ-Ensemble consisting of several Q-Learning modules to explain hyperbolic discounting in humans. Each module has its own state-action value function with different γ spanning from 0 to 1. By computing the summation of all the state-action value functions, we can approximate the hyperbolic discounting values for goal-only reward Markov decision process where non-zero rewards are only given for the goal states. However, hyperbolic discounting is different from average reward, and its decision is sometimes inconsistent. Then, we propose an alternative framework called an independent γ-Ensemble where each module learns independently by Q-learning. We show that the independent γ-Ensemble method can compute the average reward for each two neighboring modules. Figure 3.3.4 shows the proposed architecture inspired from Uchibe and Doya, (2004).
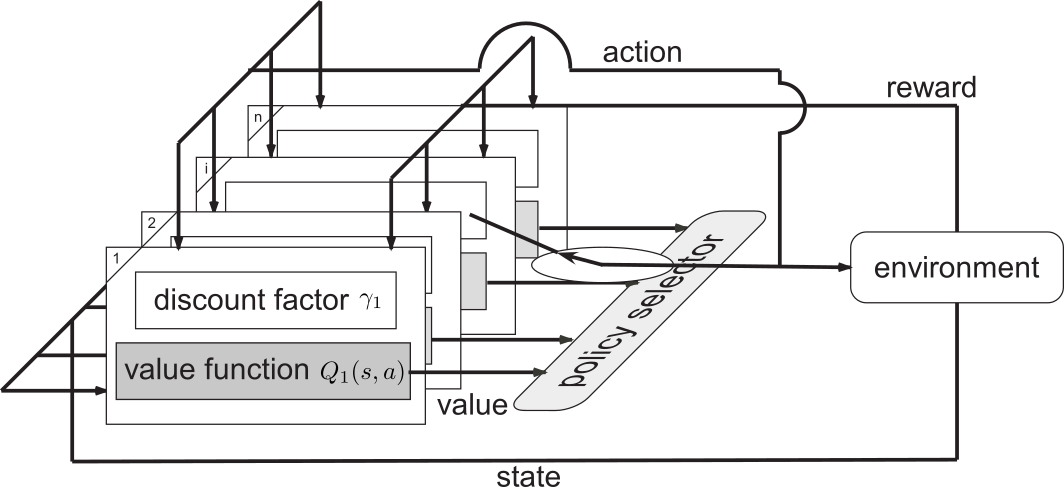
Figure 3.3.4: Architecture of the γ-Ensembles.
3.3.5 Emergence of communication between two RL agents [Huang]
Although communication is a vital activity in the development of information exchange, the means by which communication might have emerged is not yet understood. We investigated the emergence of communication between two reinforcement learning agents (Sato, Uchibe, and Doya 2008), but each agent adopts the standard Q-learning algorithm to learn an optimal policy. Consequently, we should consider a joint action of motion and communication. Figure 3.3.5 shows the proposed architecture in which each agent two Q functions: one is for motion and the other is for communication. This decomposition allows us to reduce the number of states.
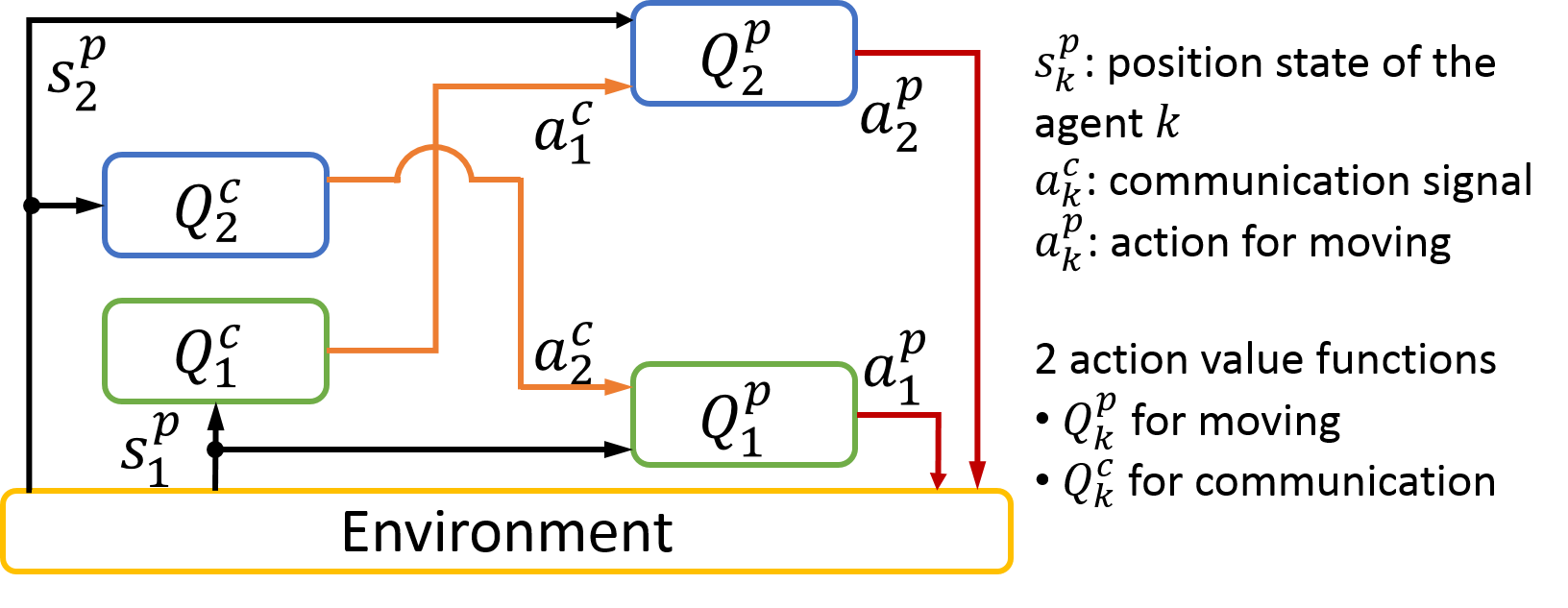
Figure 3.3.5: Multi-agent reinforcement learning framework.
We selected the "entering the same room" task with an n by n grid world, inspired from the previous study (Morita, Konno, and Hashimoto, 2012). In addition, we found that the agents could generate the meaning of signaling to represent the distance between the goal and their own position states to achieve a better performance via corporation. This study will be appeared in ICDL-EpiRob 2016.
4. Publications
4.1 Journals
- Balleine, B., Dezfouli, A., Ito, M. & Doya, K. Hierarchical control of goal-directed action in the cortical–basal ganglia network. Science Direct 5, 1-7, doi:doi:10.1016/j.cobeha.2015.06.001 (2015).
- Ito, M. & Doya, K. Parallel representation of value-based and finite state-based strategies in the ventral and dorsal striatum. doi:10.1371/journal.pcbi.1004540 (2015).
- Shimizu, Y., Yoshimoto, J., Toki, S., Takamura, M., Yoshimura, S., Okamoto, Y., Yamawaki, S. & Doya, K. Toward probabilistic diagnosis and understanding of depression based on functional MRI data analysis with logistic group LASSO. PLoS One 10, e0123524, doi:10.1371/journal.pone.0123524 (2015).
- Caligiore, D., Pezzulo, G., Baldassarre, G., Bostan, A. C., Strick, P. L., Doya, K., Helmich, R. C., Dirkx, M., Houk, J., Jörntell, H., Lago-Rodriguez, A., Galea, J. M., Miall, R. C., Popa, T., Kishore, A., Verschure, P. F. M. J., Zucca, R. & Herreros, I. Consensus Paper: Towards a systems-level view of cerebellar function: the interplay between cerebellum, basal ganglia, and cortex. doi:10.1007/s12311-016-0763-3 (2016).
- Wang, J., Uchibe, E. & Doya, K. EM-based policy hyper parameter exploration: application to standing and balancing of a two-wheeled smartphone robot. Artificial Life and Robotics 21, 125-131, doi:10.1007/s10015-015-0260-7 (2016).
- Igarashi, J., Shono, O., Moren, J., Yoshimoto, J. & Doya, K. A spiking neural network model of the basal ganglia-thalamocortical circuit toward understanding of motor symptoms of Parkinson disease, in The Brain and Neural Networks Vol. 22, 103-111 (2015).
- Moren, J., Sugimoto, N. & Doya, K. Real-time utilization of system-scale neuroscience modelsin the brain and neural networks Vol. 22 (2015).
- Uchibe, E. Forward and inverse reinforcement learning by linearly solvable Markov decision processes, in The Brain & Neural Networks Vol. 23, 2-13 (2016).
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- Baek, J., Oba, S., Yoshimoto, J., Doya, k. & Ishii, S. Presentation computational complexity reduction for functional connectivity estimation in large scale neural network, in 21st Workshop on Infomation-Based Induction Sciences and Machine Learning, IEICE, Onna, Okinawa, Japan (2015).
- Doya, K. Toward neurobiology of mental simulation, Earth-Life Science Institute Tokyo Institute of Technology(Tokyo) (2015).
- Doya, K. Neural implementation of mental simulation, in The 5th International conference on Cognitive Neurodynamics, Sanya, China (2015).
- Doya, K. Introduction to numerical methods for ordinary and partial differential equations, in Okinawa Computational Neuroscience Course 2015(OCNC2015), OIST Seaside House (2015).
- Doya, K. Introduction to reinforcement learning and Bayesian inference, in Okinawa Computational Neuroscience Course 2015(OCNC2015), OIST Seaside House (2015).
- Doya, K. Brain science and machine learning, in Consortium for Applied Neuroscience(CAN)2015 Kick-off Special Briefing Session, Chiyoda-ku, Tokyo (2015).
- Doya, K. A full-scale circuit model of the brain – exploration of the mechanisms of action control, in 19th NINS Symposium, Nagoya (2015).
- Doya, K. Neural mechanisms of reinforcement learning and decision making, University of Tokyo (2015).
- Doya, K. Neural implementation of model-based inference and planning, KAIST (Korea) (2015).
- Doya, K. Neuronal mechanism of reward-based learning and decision, in Brain Conference 2015; Joint Conference of KSBNS and KSND Korea Brain Research Institute, Daegu, KOREA (2015).
- Doya, K. Computational approaches to adaptive mechanisms of the brain, Kyungpook National University, Daegu, KOREA (2015).
- Doya, K. Presentation control of multiple representations and algorithms for action learning, in Constructive Developmental Science Meeting, Osaka University (2015).
- Doya, K. Learning algorithms and the brain architecture; Bayesian inference and mental simulation, in ISSA Summer School, Kobe (2015).
- Doya, K. Presentation The brain mechanisms for model-based predictions, in 79th Annual Convention of the Japanese Psychological Association, Nagoya (2015).
- Doya, K. Neural mechanisms of reinforcement learning and decision making, Kyoto University (2015).
- Doya, K. Artificial Intelligence and brain science, in International Conference on Intelligent Informatics and Biomedical Sciences2015, Okinawa Institute of Science and Technology, Okinawa, Japan (2015).
- Doya, K. Neural mechanisms of decision making and mental simulation, in RIKEN BSI Retreat, RIKEN Brain Science Institute, Wako, Saitama (2015).
- Doya, K. Can an integrated model of the basal ganglia-thalamocortico-spinal network and the musculoskeletal system reproduce healthy and pathological motor behaviors?, in Supercomputational Life Science 2015 (SCLS2015), Takeda Hall, The university of Tokyo, Tokyo, Japan (2015).
- Doya, K. Neural circuits and neuromodulations prediction and decision under uncertainty, in 7th International Symposium on Optogenetics:Neural Circuits and Neuromodulations, Akio Suzuki Memorial Hall, Tokyo Medical and Dental University,Tokyo, Japan (2015).
- Doya, K. Strategic research on innovative areas “Prediction and Decision Making” in The 10th Research Area Meeting "Elucidation of the Neural Computation for Prediction and Decision Making" Hitotsubashi Hall, Hitotsubashi University Tokyo, Japan (2015).
- Doya, K. Reinforcement learning by robots and the brain, cafe & studio GOBLIN.PARK, Tokyo, Japan (2015).
- Doya, K., Ishii, S. & Yamaguchi, y. Brain/MINDS - A new program for comprehensive analyses of the brain, in Neuroscience2015-38th Annual Meeting of the Japan Neuroscience Society, Kobe (2015).
- Doya, K., Magrans de Abril, I. & Yoshimoto, J. Inference of neural circuit connectivity form High-dimensional activity recording data: A Survey in The Twenty-ninth Annual Conference on Neural Information Processing Systems (NIPS2015), Montreal Convention Centre, Montreal, Canada (2015).
- Doya, K., Miyazaki, K. & Katsuhiko, M. Reward, decision making and serotonin, in 17th Joint Conference on Neurogastroenterology, Okinawa Institute of Science and Technology, Okinawa, Japan (2015).
- Doya, K., Miyazaki, K. W. & Miyazaki, K. Serotonin and the regulation of patience, in Neuroscience2015-38th Annual Meeting of the Japan Neuroscience Society, Kobe (2015).
- Eppler, J. M., Moren, J. & Djurfeldt, M. Interfaces in computational neuroscience software: Combined use of the tools NEST, CSA and MUSIC, CNS 2015, Prague (2015).
- Funamizu, A. Neural implementation of model-based decision making in posterior parietal cortex, in The 54th annual conference of Japanese Society for Medical and Biological Engineering, Nagoya (2015).
- 28. Funamizu, A. Neural substrate of Bayesian dynamic filter in posterior parietal cortex, in International Symposium on Prediction and Decision Making 2015, The University of Tokyo, Japan (2015).
- Funamizu, A. Implementation of Bayesian filter in posterior parietal cortex, in Comprehensive Brain Science Network Winter Workshop2015, Hitotsubashi Hall, Hitotsubashi University Tokyo, Japan (2015).
- Funamizu, A. Action-dependent state prediction in mouse posterior parietal cortex during an auditory virtual navigation task, in Society for Neuroscience, 2015, Chicago USA (2015).
- Funamizu, A. Neural substrate of Bayesian dynamic filter in posterior parietal cortex, in International Symposium on Prediction and Decision Making 2015, The University of Tokyo, Japan (2015).
- Funamizu, A. Action-dependent state prediction in mouse posterior parietal cortex, in International Conference on Intelligent Informatics and Biomedical Sciences2015, Okinawa Institute of Science and Technology, Okinawa, Japan (2015).
- Funamizu, A. Presentation Neural substrate of Bayesian dynamic filter in posterior parietal cortex, in Comprehensive Brain Science Network Winter Workshop2015, Hitotsubashi Hall, Tokyo, Japan (2015).
- Funamizu, A., Kuhn, B. & Doya, K. Investigating neural implementation of model-based decision making by two-photon microscopy, in The 6th FAONS Congress and 11th Biennial Conference of the Chinese Neuroscience Society (CNS), Wuzhen, CHINA (2015).
- Funamizu, A., Kuhn, B. & Doya, K. Action-dependent state prediction in mouse parietal cortex during an auditory virtual navigation task, in Neuroscience2015-38th Annual Meeting of the Japan Neuroscience Society, Kobe, Hyogo, Japan (2015).
- Gutierrez, C. E., Doya, K. & Yoshimoto, J. Self-Consistent Neuronal Population under Spike Inputs and Unbalanced Conditions, in International Conference on Intelligent Informatics and Biomedical Sciences2015, Okinawa Institute of Science and Technology, Okinawa, Japan (2015).
- Igarashi, J. A large-scale model of the basal ganglia-thalamocortical circuit toward understanding Parkinson's motor symptoms, in Japan-EU Workshop on Neurorobotics, The University of Tokyo, Bunkyo-ku, Tokyo (2015).
- Igarashi, J. Reproduction of abnormal oscillation underlying parkisonian tremor in a model of thalamocortical circuit, in 7th Acceleration Research Meeting, The Graduate School for the Creation of New Photonics Industries. Hamamatsu-shi, Shizuoka (2015).
- Igarashi, J. A large-scale model simulation of basal-ganglia-thalamocortical circuit with spinal cord and musculoskeleton on K computer: Toward understanding of information processing for motor behaviour and Parkinson's disease motor symptoms, in 25th Annual Conference of Japanese Neural Network Society (JNNS 2015), University of Electro-Communications, Chofu, Tokyo (2015).
- Igarashi, J. Anti-phase oscillations underling Parkinson's disease resting tremor in a realistic neural network model of thalamocortical circuit., in Supercomputational Life Science 2015 (SCLS2015), Takeda Hall, The university of Tokyo, Tokyo, Japan (2015).
- Ito, M. Uncovering representations and algorithms of decision making by model-based analysis of striatal neuron activities, in The 6th FAONS Congress and 11th Biennial Conference of the Chinese Neuroscience Society (CNS) Wuzhen, CHINA (2015).
- Ito, M. & Doya, K. Neural representations of memory-based and value-based decision strategies in the cortico-basal ganglia loops, in Neuroscience 2015-38th Annual Meeting of the Japan Neuroscience Society, Kobe, Hyogo (2015).
- Kannon, T., Yoshimoto, J., Amano, M., Nishioka, T., Usui, S. & Kaibuchi, K. Kinase-Associated Neural Phospho-Signaling Database: XooNIps based Neuroinformatics Database of Protein Phosphorylation, in Advances in Neuroinformatics (AINI) 2015, The University of Tokyo, Tokyo, Japan (2015).
- Kasahara, K., Charles, D. S., Honda, M. & Hanakawa, T. An inter-individual variability in peak frequency of event-related desynchronization of sensorimotor rhythms during electroencephalographic-based brain machine interface, in Neuroscience 2015-38th Annual Meeting of the Japan Neuroscience Society, Kobe, Hyogo, Japan (2015).
- Kosuke, Y., Shimizu, Y., Yoshimoto, J., Toki, S., Okada, G., Takamura, M., Okamoto, Y., Yamawaki, S. & Doya, K. Resting state functional connectivity explains individual scores of multiple clinical measures for major depression, in IEEE International Conference on Bioinformatics and Biomedicine2015, Washington D.C. USA (2015).
- Makoto, I., Tomohiko, Y. & Doya, K. Availability of working memory affects rats’ choice strategy and neuronal representation in the cortex and the striatum in The 45th Annual Meeting of the Society for Neuroscience, McCormick Place, Chicago, USA (2015).
- Makoto, I., Tomohiko, Y. & Doya, K. Working-memory availability changes rats’ choice strategy and neuronal representation in the cortex and the striatum, in International Symposium on Prediction and Decision Making 2015, The University of Tokyo, Japan (2015).
- Makoto, I., Tomohiko, Y. & Doya, K. Presentation calcium imaging of hippocampal CA1 neurons of freely-moving rats in Comprehensive Brain Science Network Winter Workshop2015, Hitotsubashi Hall, Hitotsubashi University Tokyo, Japan (2015).
- Moren, J., Igarashi, J., Yoshimoto, J. & Doya, K. A full rat-scale model of the basal ganglia and thalamocortical network to reproduce Parkinsonian tremor, in CNS 2015, Prague (2015).
- Reinke, C., Uchibe, E. & Doya, K. Maximizing the average reward in episodic reinforcement learning tasks with an ensemble of Q-Learners, in International Symposium on Prediction and Decision Making 2015, The University of Tokyo, Japan (2015).
- Reinke, C., Uchibe, E. & Doya, K. Maximizing the average reward in episodic reinforcement learning tasks, in International Conference on Intelligent Informatics and BioMedical Science2015, Okinawa Institute of Science and Technology, Okinawa, Japan (2015).
- Tokuda, T., Yoshimoto, J., Shimizu, Y. & Doya, K. Multiple clustering based on co-clustering views, in ICML 2015 workshop on features and structures, Lille, France (2015).
- Tokuda, T., Yoshimoto, J., Shimizu, Y. & Doya, k. Multiple clustering based on co-clustering views, in IJCNN 2015 workshop on Advances in Learning from/with Multiple Learners, Killarney, Ireland (2015).
- Tokuda, T., Yoshimoto, J., Shimizu, Y. & Doya, K. Multiple co-clustering based on nonparametric mixture models with heterogeneous marginal distributions, in International Meeting on High-dimensional data drive science, 2015, Mielparque Kyoto, Kyoto, Japan (2015).
- Uchibe, E. How do we define the rewards in reinforcement learning?, in Neuro Computing meeting, OIST, Okinawa (2015).
- Uchibe, E. & Doya, K. Inverse Reinforcement Learning with Density Ratio Estimation, in The Multi-disciplinary Conference on Reinforcement Learning and Decision Making 2015(RLDM2015), University of Alberta, Edmonton, Canada (2015).
- Uchibe, E. & Doya, K. Inverse reinforcement learning for behavior analysis and control, in International Symposium on Prediction and Decision Making 2015, Koshiba hall, University of Tokyo, Tokyo, Japan (2015).
- Uchibe, E. & Doya, K. Presentation Forward and inverse reinforcement learning for playing games, in CBSN Workshop 2015, Hitotsubashi hall, Hitotsubashi University, Tokyo, Japan (2015).
- Yoshimoto, J. Computational approach to understanding dopaminergic actions on striatal medium spiny neurons, in The 6th Joint Workshop on Neuroscience and Structural Biology, Okazaki Confence Center, Okazaki, Aichi, Japan (2015).
- Yoshimoto, J., Kannon, T., Amano, M., Nishioka, T., Usui, S. & Kaibuchi, K. Neural PhosphoSignaling Database: A neuroinformatics platform for protein phosphorylation, in Neuroscience 2015-38th Annual Meeting of the Japan Neuroscience Society, Kobe, Hyogo, Japan (2015).
- Yoshimoto, J., Kannon, T., Amano, M., Nishioka, T., Usui, S. & Kaibuchi, K. Neural PhosphoSignaling Database: A neuroinformatics platform for protein phosphorylation with quality control, in Neuroinformatics 2015, Cairns, Australia (2015).
- Yoshimoto, J., Yukinawa, N. & Nakano, T. Computational modeling of dopaminergic actions on striatal medium spiny neurons, in Symposium on Principle and breakdown of higher brain function by decoding intracellular signaling in the 58th Annual Meeting of the Japanese Society for Neurochemistry, Omiya, Saitama, Japan (2015).
- Yoshizawa, T., Ito, M. & Doya, K. The role of the cortico-basal ganglia loops for reward-motivated choice motor behavior, in Neuroscience2015- 38th Annual Meeting of the Japan Neuroscience Society Kobe, Hyogo, Japan (2015).
- Doya, K. Neural mechanisms of mental simulation, Peking University, Beijing, China (2016).
- Doya, K. What can we do with whole brain connectivity and activity data?, in AMED-NIH brain workshop, AMED, Chiyoda-ku, Tokyo, Japan (2016).
- Doya, K. Neural mechanisms of mental simulation, in NIPS Symposium "Perspectives in Next-Stage Functional Life Science", Okazaki Cnference Center, Okazaki, Aichi, Japan (2016).
- Doya, K. Brain science in the age of high-throughput measurements and high-performance computing, Kobe University, Kobe, Hyogo, Japan (2016).
- Funamizu, A. Investigation of hierarchical computation in cortical column by two-photon microscopy, in Symposium for Shingakujyutsu, sparse modeling, Kobe university, Kobe, Hyogo, Japan (2016).
- Funamizu, A. Neural implementation of Bayesian inferece ~investigating with theretical and experimental approaches~, NAIST, Nara, Japan (2016).
- Funamizu, A. Neural substrate of dynamic Bayesian inference in the cerebral cortex., Cold Spring Harbor Laboratory, New York, US (2016).
- Funamizu, A. Neural substrate of dynamic Bayesian inference in the cerebral cortex, in International Symposium on Adaptive Circuit Shift, Doshisya University, Kyoto, Japan (2016).
- Funamizu, A. Presentation Investigation of hierarchical computation in cortical column by two-photon microscopy, in Symposium for Shingakujyutsu, sparse modeling, Kobe university, Kobe, Hyogo, Japan (2016).
- Funamizu, A. Neural substrate of dynamic Bayesian inference in the posterior parietal cortex., in The Winter Workshop 2016 on Mechanism of Brain and Mind, Rusutsu, Hokkaido, Japan (2016).
- Hamada, H. Analysis of intrinsic functional networks of the mouse brain and its application, Tohoku University, Sendai, Miyagi, Japan (2016).
- Hamada, H., Sakai, Y., Hikishima, K., Takata, N., Tanaka, K. & Doya, K. Intrinsic functional networks in awake mice, in The 16th Winter Workshop on the Mechanism of Brain and Mind, Rusutsu, Hokkaido, Japan (2016).
- Igarashi, J., Moren, J., Yoshimoto, J. & Doya, K. Lateral inhibition and movement selection of a three dimensional model of mouse primary motor cortex in "National Institute for Physiological Science Symposium 2016 Prospect of functional life science in the next stage", Okazaki Cnference Center, Okazaki, Aichi, Japan (2016).
- Makoto, I. & Doya, K. Information coded in hippocapal CA1: Calcium imaging from freely-moving rats, in The Third CiNet Conference -Neural mechanisms of decision making: Achievements and new directions, CiNet Bldg. Suita, Osaka, Japan (2016).
- Miyazaki, K. The role of serotonin in the regulation of waiting behavior for future rewards, in The Winter Workshop 2016 on Mechanism of Brain and Mind, Rusutsu, Hokkaido, Japan (2016).
- Miyazaki, K. Neural computation mechanism of prediction and decision making by dorsal raphe serotonin neurons, in The 89th Annual Meeting of the Japanease Phaemaciligical Society, Pacifico Yokohama, Yokohama, Kanagawa, Japan (2016).
- Reinke, C. Research opportunities at the Okinawa Institute of Science and Technology & Brain inspired decision making algorithms, Carl von Ossietzky Universität, Oldenburg, Germany (2016).
- Reinke, C., Uchibe, E. & Doya, K. From neuroscience to artificial intelligence: maximizing average reward in episodic reinforcement learning tasks with an ensemble of Q- Learners, in The Third CiNet Conference: Neural mechanisms of decision making: Achievements and new directions, CiNet Bldg. Suita, Osaka, Japan (2016).
- Reinke, C., Uchibe, E. & Doya, K. Learning of stress adaptive habits with an ensemble of Q-Learners, in The 2nd International Workshop on Cognitive Neuroscience Robotics, Sankei Conference Osaka Umeda, Osaka, Japan (2016).
- Uchibe, E. Basics and recent advances in reinforcement learning, Fujitsu Laboratories, Kawasaki, Kanagawa (2016).
- Yoshimoto, J., Yukinawa, N. & Nakano, T. An informatics study on dopaminergic actions on striatal medium spiny neurons, in The Winter Workshop 2016 on Mechanism of Brain and Mind, Rusutsu, Hokkaido, Japan (2016).
- Yoshizawa, T., Ito, M. & Doya, K. The activities of striatal patch neurons in classical conditioning, in The 16th Winter Workshop on the Mechanism of Brain and Mind, Rusutsu, Hokkaido, Japan (2016).
- Yukinawa, N., Doya, K. & Yoshimoto, J. In silico screening of ion channels for dopamine-dependent excitability modulation of striatal medium spiny neurons, in SIG BIO, IPSJ, JAIST, Nomi, Ishikawa, Japan (2016).
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 Seminar
A computational approach to control memory states in biologically plausible neural networks and its application to dementia
- Date: May 7, 2015
- Venue: OIST Campus Lab1
- Speaker: Dr. Hiromichi Tsukada (Hokkaido University)
Current Activity for Open Innovation by CAN:Consortium for Applied Neuroscience
- Date: May 19, 2015
- Venue: OIST Campus Lab1
- Speaker: Dr. Ippei Hagiwara (NTT data)
DECODE Project : Connecting Omics and Imaging
- Date: June 1, 2015
- Venue: OIST Campus Lab1
- Speaker: Dr. Makoto Taiji (Quantitative Biology Center)
The dentate-hippocampal network support spatial working memory.
- Date: November 4, 2015
- Venue: OIST Campus Lab1
- Speaker: Dr. Takuya Sasaki (The University of Tokyo)
Dynamics of perceptual decision-making under temporal uncertainty.
- Date: November 25, 2015
- Venue: OIST Campus Lab1
- Speaker: Dr. Nestor Parga (Universidad Autonoma de Madrid)
Dynamic Boltzmann machines
- Date: March 28, 2016
- Venue: OIST Campus Center Bldg
- Speaker: Dr. Takayuki Osogami (IBM Academy of Technology, IBM Research - Tokyo)
6.2 Sponsored Research Meeting: The 9th Research Area Meeting of the Elucidation of the Neural Computation for Prediction and Decision Making
- Date: April 25-27, 2015
- Venue: OIST Seaside House
- Organizer: Grant-in Aid for Scientific Research on Innovative Areas, MEXT, JAPAN, Elucidation of the Neural Computation for Prediction and Decision Making
- Speakers:
- Dr. Bernd Kuhn (OIST)
- Dr. Makio Torigoe (RIKEN)
- Dr. Ichiro Maruyama (OIST)
6.3 Joint Workshop: Neuro-Computing, Bioinformatics, Mathematical modeling and Machine Learning
- Date: June 23-25, 2015
- Venue: OIST Capmpus, Semiar Room B250 & Seminar Room C210
- Co-organizers:
- The Institute of ElectronicsInformation and Communication Engineers(IEICE)
- Information Processing Society of Japan
- IEEE Computational Intelligence Society Japan Chapter
- Japan Neural Network Society
- Speakers:
- Dr. Eiji Uchibe (OIST)
- Dr. Kunihiko Fukushima (Fuzzy Logic Systems Institute)
- Dr. Kozo Kaibuchi (Nagoya University)
6.4 Sponsored Research Meeting: International Symposium on Prediction and Decision Making 2015
- Date: October 31- November 1, 2015
- Venue: Koshiba Hall, The University of Tokyo
- Speakers:
- Dr. Robert Malenka (Stanford University)
- Dr. Anatol Kreitzer (Gladstone Institutes / Universi ty of California San Francisco)
- Dr. Bernard Balleine (University of Sydney)
- Dr. Catharine A. Winstanley (University of British Columbia)
- Dr. Katsuyuki Kaneda (Kanazawa University)
- Dr. Takayuki Teramoto (Kyushu University)
- Dr. Toshio Yamagishi (Hitotsubashi University)
- Dr. John Philip O’Doherty (California Institute of Technology)
- Dr. Masahiko Haruno (NICT Cinet)
- Dr. Satoshi Umeda (Keio University)
- Dr. Takafumi Minamimoto (National Institute of Radiological Sciences)
- Dr. Hitoshi Okamoto (RIKEN Brain Science Institute)
- Dr. Wolfram Schultz (University of Cambridge)
- Dr. Nathaniel Daw (New York University)
- Dr. Masamichi Sakagami (Tamagawa University)
- Dr. David Redish (University of Minnesota)
- Dr. David Foster (The Johns Hopkins University)
- Dr. Sophie Deneve (CNRS)
- Dr. Akihiro Funamizu (OIST)
- Dr. Yu Ohmura (Hokkaido University)
- Dr. Akio Namiki (Distinguished McKnight University / University of Minnesota)
- Dr. P. Read Montague (University College London / Virginia Tech)
- Dr. Mitsuo Kawato (ATR)
- Dr. Mathias Pessiglione (CM Brain & Spine Institute)
- Dr. Michael J. Frank (Brown University)
- Dr. Hidehiko Takahashi (Kyoto University)
7. Other
Nothing to report.



