FY2020
Biological Systems Unit
Professor Igor Goryanin
Abstract
For last years, the Biological Systems Unit has been engaged in development of BioElectrochemical System (BES)/Microbial Fuel Cell (MFC) technology for wastewater treatment. The BES/MFC applies complex interactions between microbial populations and electrodes to remove organics and to generate electricity, merging basic and applied goals of our Unit. By utilizing biological, chemical, engineering, and bioinformatics approaches, the Unit seeks to improve BES/MFC systems for better treatment efficiencies and electricity generation by understanding and building ideal microbial communities and developing cost-effective materials. Sustainable wastewater treatment is especially important for small islands like Okinawa.
One of the challenges of this technology is scaling up. The Unit has been working with a local Awamori (Okinawa unique spirit) distillery for the past eight years to treat rice wash and distillery wastewater. David Simpson received Tech Pioneer grant from OIST Business Development Section and continued his challenges to treat Awamori rice and distillery wastes for mid-and small factories with newly developed reactors. The Unit continued with technology development for in-house anion exchange membrane under OIST POC II project to be utilize for cost down for the scale up system. The results are protected by patents (Separator of Microbial Fuel Cells and Phosphate removal system).
With another POCII project, we aimed to selectively treat and recycle distillery and bioethanol fermentation waste using MFC by removing volatile fatty acids VFAs, by product of fermentation, which is inhibitory for recycling distillery waste. The project was then directed to develop a biosensor for monitoring VFA in fermentation waste. Also a unique high-throughput screening and selection system of PCB-bioelectrocholeaching was developed using 96-well plate array.
Lastly, the Unit continued developing organics, odor, nitrogen, and phosphate removal technology from raw and aerated swine wastewater using biocathode system. The demonstration reactors were monitored at the Okinawa Prefecture Glassland and Livestock Research Center and improvements were made. We focus on the scaling up of the technology and sleeking for the additional funding and collaborations.
1. Staff
- Dr Igor Goryanin, Professor
- Dr. Mami Kainuma, Group Leader
- Dr. Peter Babiak, Staff Scientist
- Dr. Anna Prokhorova, Postdoctoral Fellow
- Dr Marsel Murzabaev, Technical Staff
- Mr. Geoffrey Schaffer-Harris, Technical Staff
- Ms. Shizuka Kuda, Research Unit Administrator
2. Collaborations
2.1 A microbial fuel cell-based system for winery wastewater reuse (USA)
- Type of collaboration: Joint research
- Researchers:
- Professor Michael F. Cohen, Sonoma State University, USA
2.2 Development of Microbial Fuel Cell-based technologies for wastewater treatment and characterization
- Type of collaboration: Joint research
- Researchers:
- Dr. Gábor Márk Tardy, Budapest University of Technology and Economics, Hungary
- Dr. Miklós Gyalai-Korpos, BES Europe Ltd. / PANNON Pro Innovations Ltd.
2.3 COVID-19 and human microbiome, analytical methods for metagenomes
- Type of collaboration: Joint research
- Researchers:
- Dr. Anatoly Sorokin / Moscow Institute of Physics and Technology
- Dr. Olga Vasieva / Ingenet Ltd.
3. Activities and Findings
3.1. Bioelectrochemical Systems (BES): Material development, applications, scaling up and microbial metagenomic.
The following projects are dedicated to improving our BES technology, including development of cost-effective bioreactor materials, widening possible applications of our technology, including scaling up and understanding microbial communities.
3.1.1 Anion exchange hydrogels development for wastewater treatment
The project focused on commercialization of membrane technology developed in Goryanin unit. Scale up of in-house developed membrane was optimized and cost-effective projection was achieved for industrial applications. Datasheets for both anion and cation exchange membrane are now available and production process was optimized.
Our in-house cation exchange membranes have been already utilized in our scale up projects including BioAlchemy (Spin-off company) for treatment of wastewater containing organic solvent, and Awamori distillery waste and rice wash wastewater. Anion exchange membranes are being tested using swine wastewater for nitrate and phosphate removal.
The technology is protected by patent WO2019160046A1 Separator of a microbial fuel cell. In addition, the phosphorus removal technology is protected under JP 2021-90950.
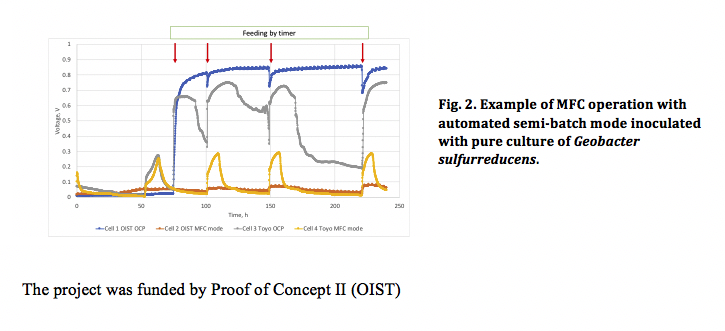
The project was funded by Proof of Concept II (OIST).
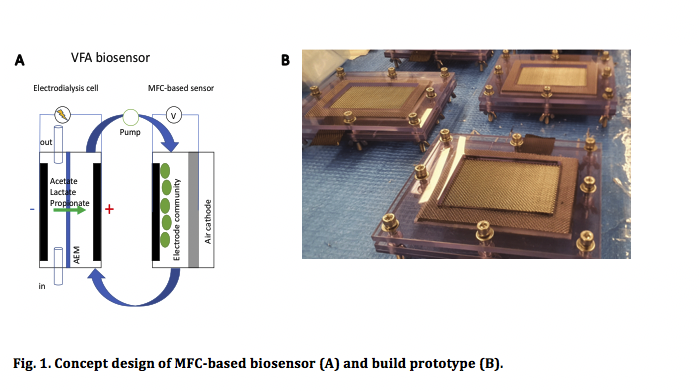
3.1.2. Development of MFC-based biosensor
Another promissing direction of this technology is developing a biosensor for volatile fatty acides (VFA) monitoring in fermentation waste. Why is VFA monitoring important? VFA are the byproducts of almost all biotech fermentation processes, including ethanol and biogas production, their high concentration may inhibit yeast activity during the late stages of fermentation at distilleries. The concentration of VFA is an established indicator for monitoring of the anaerobic process. It gives fast and reliable information of process status compared to other common indicators such as pH, alkalinity, gas production and gas composition. Monitoring VFA in fermentation mixture give us the ability to stop fermentation at the best time for highest yield of production (i.e. ethanol). All currently available VFA detection systems are based on filtration, with consequent fouling problems.
We developed a prototype of a MFC-based biosensor, which does not require filtration, was designed and tested with a synthetic wastewater. The system comprised of two-steps: 1) an electrodialysis reactor with two chambers separated by our proprietary anion exchange membrane and 2) a MFC-based biosensor.
By applying potential, VFA is removed from one chamber and concentrated in the other chamber. Then the VFA concentrated wastewater is pumped through the MFC where VFA will be consumed and converted to electricity by specially evolve and augmented electrogenic bacterial community. The electrodialysis VFA separation device is scalable in principle and can be used for VFA removal as well. The amount of energy spent in separation stage will be combined with amount of energy produced in MFC. These parameters will be used to calibrate the sensor for VFA monitoring.
3.1.3. Metabolic engineering of a novel strain of electrogenic bacterium Arcobacter butzleri to create a platform for single analyte detection using a microbial fuel cell
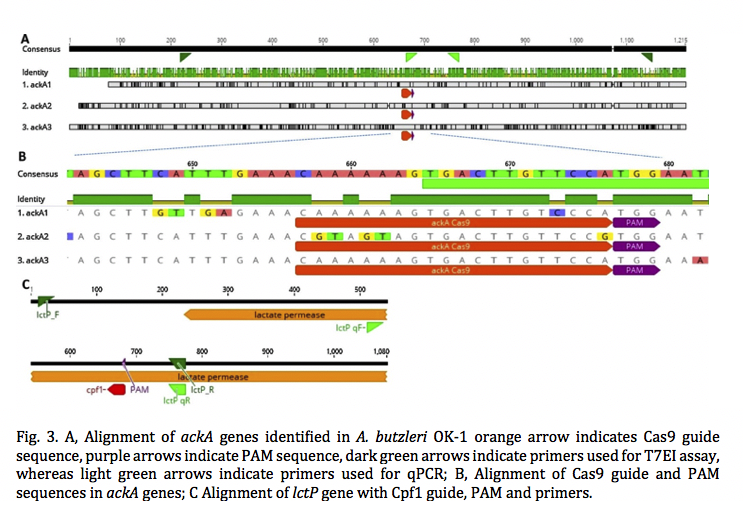
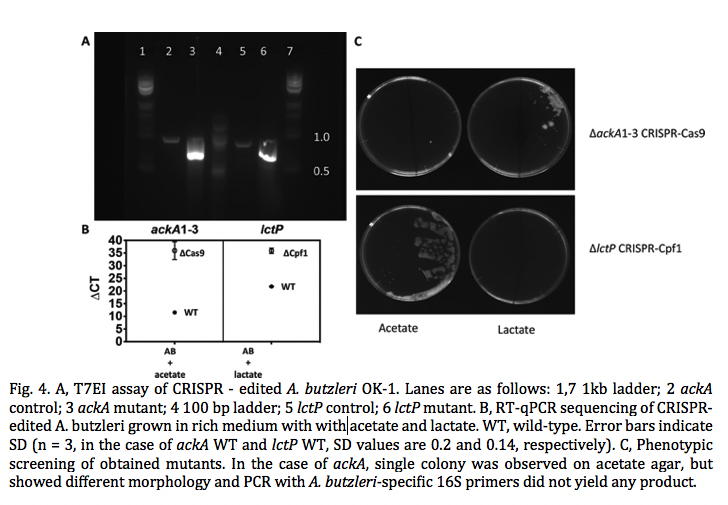
Electrogenic bacteria metabolize organic substrates by transferring electrons to the external electrode, with subsequent electricity generation. In this proof-of-concept study, we present a novel strain of a known, electrogenic Arcobacter butzleri that can grow primarily on acetate and lactate and its electric current density is positively correlated (R2 = 0.95) to the COD concentrations up to 200 ppm. Using CRISPR-Cas9 and Cpf1, we engineered knockout A. butzleri mutants in either the acetate or lactate metabolic pathway, limiting their energy metabolism to a single carbon source. After genome editing, the expression of either acetate kinase, ackA, or lactate permease, lctP, was inhibited, as indicated by qPCR results. All mutants retain electrogenic activity when inoculated into a microbial fuel cell, yielding average current densities of 81–82 mA/m2 , with wild type controls reaching 85–87 mA2 . In the case of mutants, however, current is only generated in the presence of the substrate for the remaining pathway. Thus, we demonstrate that it is possible to obtain electric signal corresponding to the specific organic compound via genome editing. The outcome of this study also indicates that the application of electrogenic bacteria can be expanded by genome engineering.
Szydlowski, L., Lan, TCT., Shibata, N., and Goryanin, I. (2020) Metabolic engineering of a novel strain of electrogenic bacterium Arcobacter butzleri to crat a platform for single analyte detection using a microbial fuel cell. Enzyme and Microbial Technology. 139: 109564 https://doi.org/10.1016/j.enzmictec.2020.109564
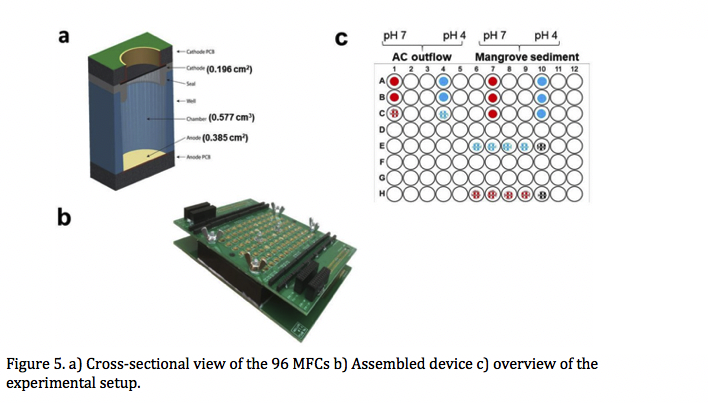
3.1.4. High-throughput screening and selection of PCB-bioelectrocholeaching, electrogenic microbial communities using single chamber microbial fuel cells based on 96-well plate array.
We demonstrate a single chamber, 96-well plate based Microbial Fuel Cells (MFCs). This invention is aimed at robust selection of electrogenic microbial community under specific conditions, (pH, external resistance, inoculum) that can be altered within the 96 well plate array. Using this device, we selected and multiplicated electrogenic microbial communities that can operate under different pH and produce current densities up to 19.6 A/m3. Moreover, studies shown that Cu mobilization through PCB bioleaching occurred, thus each communities were able to withstand presence of copper ions up to 600 mg/L. Metagenome analysis reveals high abundance of Dietzia spp., previously characterized in MFCs, but not reported to grow at pH 4, as well as novel species, not yet associated with electrogenicity, e.g. Microbacterium spp. Microscopic observations (combined SEM and EDS) reveal that some of the species present in the anodic biofilm were accumulating copper on their surface, probably due to the presence of metalloprotein complexes on their outer membranes. Taxonomy analysis indicated that similar consortia populate anodes, cathodes and OCP controls, although total abundances of aforementioned species are different among those groups. Annotated genomes showed high presence of multicopper oxidases and Cu-resistance genes such as cupredoxins and copCD. Although all inocula contained some known electrogenic taxa, enriched microbial communities were derived only from one inoculum (AC outflow). We thus conclude that electrochemically-driven bioleaching of PCB created conditions that selectively enriched these groups.
Szydlowski, L., Ehlich, J., Shibata, N., Goryanin, I. High-throughput screening and selection of PCB-bioelectrocholeaching, electrogenic microbial communities using single chamber microbial fuel cells based on 96-well plate array. BioRxiv. doi: https://doi.org/10.1101/2021.06.09.447729 (in press)
3.1.5. R&D on swine wastewater treatment technology for simultaneous removal of nitrate and organics using bioelectrochemical systems (BES)
The volume of wastewater produced by intensive pig farming in Okinawa surpasses the available capacity for treatment and recycling. Removal of nitrate from wastewater is a worldwide concern due to its negative effects on human and environmental health. Currently the nitrate-nitrogen discharge limit for livestock wastewater receives special measure (500 mg/L) but very near future it will be lower to the general discharge standard (100 mg/L) to meet all other industries, and cost-effective nitrate-nitrogen removal in livestock industry is the urgent issue in Japan.
Since May, 2019, the pilot-scale biocathode reactors (58 L active volume) have been running at Okinawa Prefecture Glassland and Livestock Research Center and monitored for over one year at continuous mode. The system simultaneously treated organics and nitrate in real swine wastewater. The full-strength raw wastewater containing high organic and volatile fatty acid levels (responsible for malodor) that are oxidized by the microbial community in the anode chamber which support lowering the aeration time and removing smells. Under applied potential, electrons are transferred to the cathode, where nitrate from aeration-treated wastewater (nitrified) serves as an electron acceptor and converted to dinitrogen by denitrification via the cathodic microbial community. The advantage of this system is the allowing of denitrification at low-COD/N wastewater. This year, the maintenance and improvements were made for the optimal operations.
The long-term microbial communities were analyzed on the electrodes at lab-scale by whole genome shotgun sequencing. The results revealed that the highest microbial diversity was detected when the cathode was poised to -0.6 V which included autotrophic denitrifier such as Syderoxidans, Gallionerla and Thiobacilllus genus. Among various bacteria, Thiobacillus denitrificans, known electrotrophic bacteriawas enriched over 60% ofall communities after 6-month operations, indicating that importance of applying potential to cathode for efficient nitrate removal. Through metatranscriptome analysis we hypothesized a possible mechanism of denitrification process with different bacteria involvement.

The project is originally funded by Okinawa Prefectural Government and will continue with POC I. conducted with a research body with Okinawa Environment Science Center, Okidoyaku and Okinawa Prefectural Grassland and Livestock Research Center.
Prokhorova, A., Kainuma, M., Hiyane, R., Boerner, S., Goryanin, I., (2020) Concurrent treatment of raw and aerated swine wastewater using an electrotrophic denitrification system., Bioresource Technology., Volume 322: https://doi.org/10.1016/j.biortech.2020.124508
3.2 Microbiome, analytical methods for metagenomics.
3.2.1. ASAR upgrade and migration completed. Please see the details below
Deigo migration report
Replacement of Sango with Deigo cause a significant change in computational infrastructure. So, it was not possible to run scripts from the home directory anymore. Those requirements disrupt the original concept of ASAR scripts and other computational infrastructure used on Sango. We have focused on ASAR scripts, as they are the most frequently used computational infrastructure, and all scripts were rewritten from scratch.
New 34 ASAR scripts required for fetching data to/from MG-RAST are available on GitHub: https://github.com/lptolik/slurm_scripts. Modified algorithm of data ASAR script invocation is created
We installed and configured Nextflow software on Deigo server and refactor YAMP metagenomic annotation pipeline to be working on Deigo.
3.2.2 Comparison of metagenomics (DNA) and metatranscriptomics (RNA) data on the example of evolving electrogenic community
We have done statistical analysis of the metagenomic and metatranscriptomics on MFC datasets. The first step of DNA/RNA sequencing data comparison was the identification of common and data specific parts of the annotation.
It could be seen that in general metagenomics and metatranscriptomics are agreed on functional annotation. The intersection of two classes contains majority of data in both abundance and annotation diagrams. On the following graph it could be seen that there is also great correlation between KO annotations abundances in both metagenomics and metatranscriptomics. Classes are well balanced between DNA and RNA data.
For genera situation is more complicated that for families, however in average genera located out of group union have less than 100 reads and would be discarded by further analysis.
The source code of ASAR software was modified to allow simultaneous visualization of both metagenomics (DNA) and metatranscriptomics (RNA) data. As in original ASAR window, there are three projections of data hypercube:
- F/T panel to analyse function distribution along taxa in particular sample;
- F/M panel to analyse function distribution in samples;
- T/M panel to analyse taxonomy abundance in samples.
As previously, the left side of window is the sidebar, which contains a set of options to determine the selection, aggregation and visualisation parameters of the projection. The right side of the page is our heat map module. The top is the DNA heatmap, and the bottom is the RNA heatmap. The design of two heat maps on the same page can greatly improve production efficiency.
F/T panel
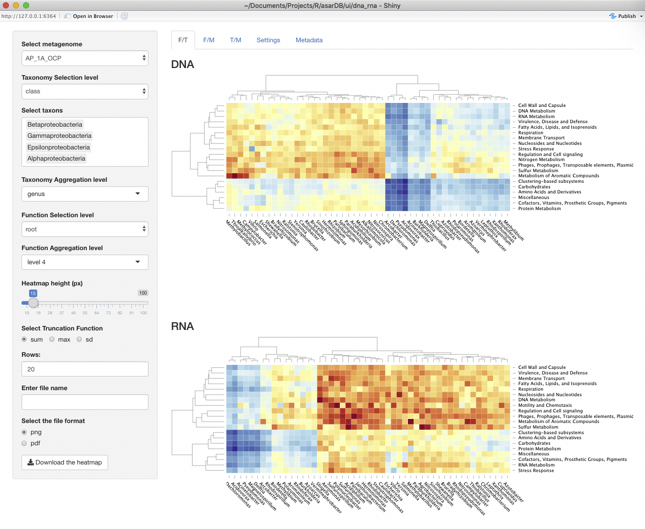
Here we show how abundances of genes providing particular function to the particular taxon are distributed within one sample. In the case of the screenshot above all functions are aggregated at the topmost level and only genes from Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria and Epsilonproteobacteria are shown. Those classes are responsible for 49% of RNA reads and 51% of DNA reads. The level of the function aggregation was chosen due to absence of non-overlapping functions at this level.
F/M panel
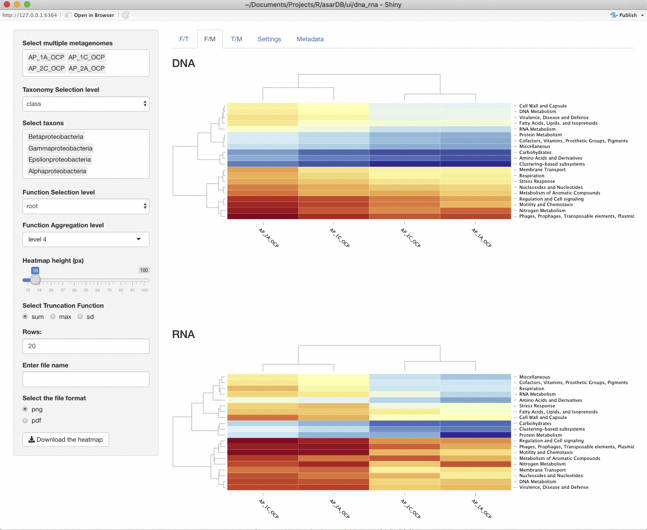
On the F/M panel we can analyse distribution of reads among functions in different samples. Here it could be seen that grouping of samples is the same in both datatypes and that in general distribution of abundances are similar. However in transcriptomics data Amino acid metabolism is clustered with respiration, while in genomics data it clustered with protein metabolism. It could be easily explained if we notice that respiration is the most distinct group between DNA and RNA data. That means that genes of that group are expressed much more actively. The opposite dependency could be observed for Cell wall metabolism which is less abundant in RNA data in comparison with DNA data.
T/M panel
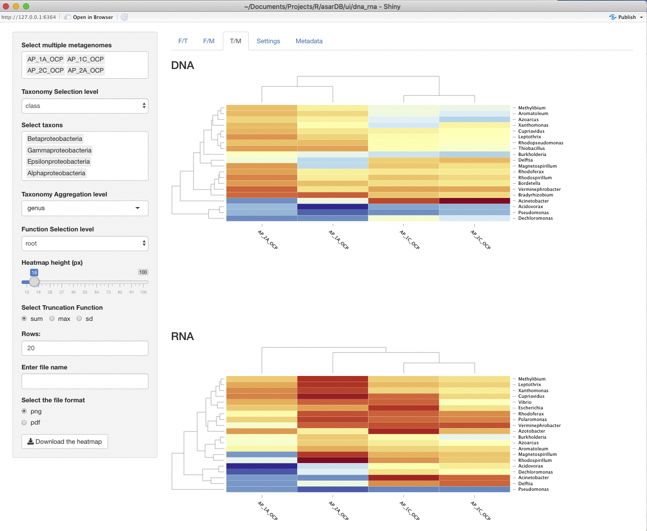
This panel let us compare taxon abundances among samples. In contrast to F/M panel samples here arranged differently, which means the activity of genera is different from its abundance. For example Acidovorax is abundant in all samples in DNA data almost evenly, while its abundance in RNA data rapidly declines between 1A and 2C.
3.2.2 Review article The Gut microbiome versus COVID-19 (O.Vasieva, I.Goryanin, 2020)
A digestive system is an environmental frontline involving digestive secretions, intestinal cell metabolism and gut microbiome that significantly modulate multiple organism’s functions. Understanding of the ’’gut-lung axes’’, where gut residential microbiota play an important role, may help to develop better prophylactics and intervention strategies for diseases caused by respiratory viruses, including COVID-19. Gastrointestinal symptoms are common in COVID-19 patients and generally indicative of COVID-19 complications. As we have learned so far, diarrhea and gut dysbiosis during SARS-CoV-2 infections should not be ignored but can be used to distinguish pathways of dysregulation of the immune system and the regulatory pathways upstream and downstream viral primary binding receptors, such as ACE2. This review presents existing evidence on microbiome signatures in the gut and respiratory system that may predict severity and long-term outcomes of COVID-19 disease. Understanding of the factors (such as a pro-inflammatory trends, metabolite availability modulation, impact on cell signaling and pathogenic properties) translating an effect of the microbiome composition to the severity of respiratory infections should help with development of new approaches to health monitoring, disease prevention and treatment.
4. Publications
4.1 Journals
- Szydlowski, L., Lan, TCT., Shibata, N., and Goryanin, I. (2020) Metabolic engineering of a novel strain of electrogenic bacterium Arcobacter butzleri to crat a platform for single analyte detection using a microbial fuel cell. Enzyme and Microbial Technology. 139: 109564 https://doi.org/10.1016/j.enzmictec.2020.109564
- Kamennaya, NA, Gray, J., Ito, S., Kainuma, M., …..Holman, HY., Torok., T., and Cohen, M. (2020) Deconstruction of plant biomass by a Cellulomonas strain isolated from an ultra-basic (lignin stripping) spring. Archives of Microbiology 202*1077-1084.
- Prokhorova, A., Kainuma, M., Hiyane, R., Boerner, S., Goryanin, I., (2020) Concurrent treatment of raw and aerated swine wastewater using an electrotrophic denitrification system., Bioresource Technology., Volume 322: https://doi.org/10.1016/j.biortech.2020.124508
- Tardy, GM., Lorant, B., Gyalai-Korpos, M., Bakos,V., Simpson, D., Goryanin. I. (2020) Microbial fuel cell biosensor for the determination of biochemical oxygen demand of wastewater samples containing readily and slowly biodegradable organics. Biotechnology Letters. 43:445-454. https://doi.org/10.1007/s10529-020-03050-5
- Lorant, B., Gyalai-Korpos, Goryanin, I., Tardy, GM. Application of air cathode microbial fuel cells for energy efficient treatment of dairy wastewater. Periodica Politechnica Chemical Engineering. 65(2)200-209. https://doi.org/10.3311/PPch.16695 (in press)
- Szydlowski, L., Ehlich, J., Shibata, N., Goryanin, I. High-throughput screening and selection of PCB-bioelectrocholeaching, electrogenic microbial communities using single chamber microbial fuel cells based on 96-well plate array. BioRxiv. doi: https://doi.org/10.1101/2021.06.09.447729 (in press)
- Vasieva, O & Goryanin, I 2021, 'The Gut Microbiome versus COVID-19', Journal of Computer Science and. Systems Biology, vol. 14, no. 1, pp.
- Cohen MF, Kubota CM, Quintero Plancarte G, Kainuma M (2021) Biological polishing of liquid and biogas effluents from wastewater treatment systems. In: Integrated and hybrid technology for water and wastewater treatment. Ang WL, Mohammad A., Eds., Elsevier, ISBN: 595806, in press
4.2 Oral and Poster Presentations
- Kainuma, M and Babiak, P. Okinawa Manufacturing Technology Exhibition 2020 Corporate Exhibition (沖縄ものづくり技術展Jan 27-28, virtual presentation)
- Kainuma, M., Prokhorova, A., Hiyane, R., Boerner, S., Kazeoka, M., Ninomiya, K. and Goryanin, I. Long-term operation of biocathodic denitrification to treat low C/N swine wastewater using electrotrophic bacteria. Livestock Waste 2020. Tsukuba, Japan. March 28, 2021 (Best Poster Award).
5. Intellectual Property Rights and Other Specific Achievements
- Fedorovich, V., Filonenko, G., Goryanin, I., Schaffer-Harris, KG., Simpson, DJW and Babiak, P., Separator of a microbial fuel cell, W2919160046A1
- Babiak, P., Schaffer-Harris, KG., Kainuma, M. and Goryanin, I. Method for treating wastewater and wastewater treatment system JP2021-90950



