FY2011 Annual Report
Biological Systems Unit
Professor Igor Goryanin

Abstract
The main focus of our research is developing the microbial fuel cells. Starting with the lab-scale systems we moved toward the practical application by scaling-up the reactors and developing systems integration.
1. Staff
- Dr. Larisa Kiseleva, Researcher
- Midori Tanahara, Administrative Assistant
2. Collaborations
- Theme: Biological network analysis
- Type of collaboration: Research Collaboration
- Researchers:
- Dr. Tong Hao, Tianjin University
3. Activities and Findings
3.1 Electrogenic microbial communities isolated from Okinawa sea sediments
This project involves both experimental work and computational analysis and relates to the innovative process of bio-electroremediation that rests on the unique physiology of microorganisms capable of conserving energy through extracellular electron transfer to solid conductive surfaces. These microorganisms are known as electricigens and used in the microbial fuel cells (MFCs), the systems that exploit microbial catabolic activities to generate electricity from a variety of biodegradable materials, including complex organic waste.
Given organic waste as an input and electrical power as output, MFC comprise an absolutely advantageous eco-friendly bio-electroremediation system. Beyond wastewater treatment MFCs have a number of potential uses, such as electrical power supplies for environmental sensors, electronic devices, and electric vehicles.
Although several microbial species are known to be capable of the conversion of organic matter into electricity, it is important to determine the structure of most successful electrogenic community for the best performance of microbial fuel cell system in given conditions.
The aim of this research is to isolate new electrogenic microorganisms from Okinawa natural environment, characterize their potential to industrial waste treatment, and study molecular mechanisms of electron transfer. At present, we have isolated a mixed bacterial community capable of electricity generation in the lab-scale MFC and having great metabolic potential of breaking down a wide range of organic and inorganic pollutants.
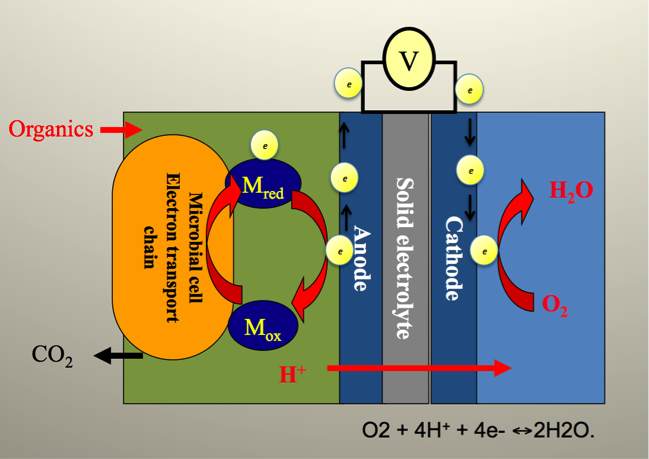
Figure 1: General principles of microbial fuel cell system. MFC provides exoelectrogenic microorganisms with a solid electron acceptor in form of anode. The electrons released by microbes are captured by anode and transferred to cathode according to the potential gradient between them. Protons released at the anode are transferred to cathode diffusively through cation exchange membrane or without. Chemical reactions occurring at cathode utilize the arrived electrons and protons as well as oxygen, thereby producing pure water and completing an electric circuit.
A marine sediment near Kaichu-Doro (Pacific coast, Okinawa island) was used as a source of microorganisms to inoculate a lab-scale MFC (Fig.2). After two month of continious operation the MFC was reassembled and microbial compositions from anodic surface and anodic chamber were studied in comparison with initial marine microbial association.
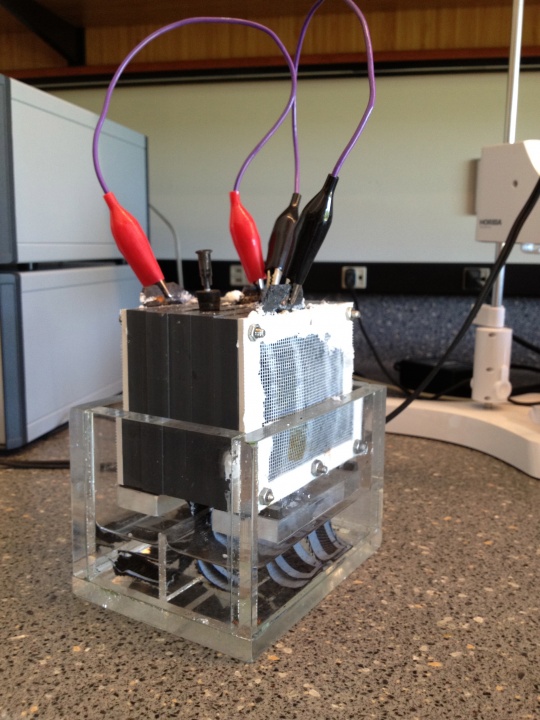
Figure 2: Lab-scale microbial fuel cell. A single chamber mediatorless MFC containing four electrodes (two anodes and two cathodes) connected by copper wires.
Scanning and transmission electron microscopy was used to visualize microorganisms attached to anode. The carbon cloth fibers of the anode were colonized by rod-shaped bacteria, and the carbon granules microbial populations predominated by helical-shaped bacteria (Fig.3). Micrograph images indicate formation of pili, presumably nanowires, by helical shaped bacteria which implies their possible electrogenic properties.
The electrically conductive appendages, or nanowires, are used by exoelectrogenic bacteria to transfer electrons to terminal electron acceptors. Nanowires are most studied for representatives of Shewanella and Geobacter genera, the rod-shaped bacteria normally abundant in the anodic biofilms. However, the group of spiral-shaped bacteria have not been studied in the context of electricity generation.
According to following genetic analyses, the helical bacteria that survived and expanded under applied selective pressure of MFC were related to genera Magnetospirillum and genera Rhodospirillum, both belonging to class of Alphaproteobacteria.
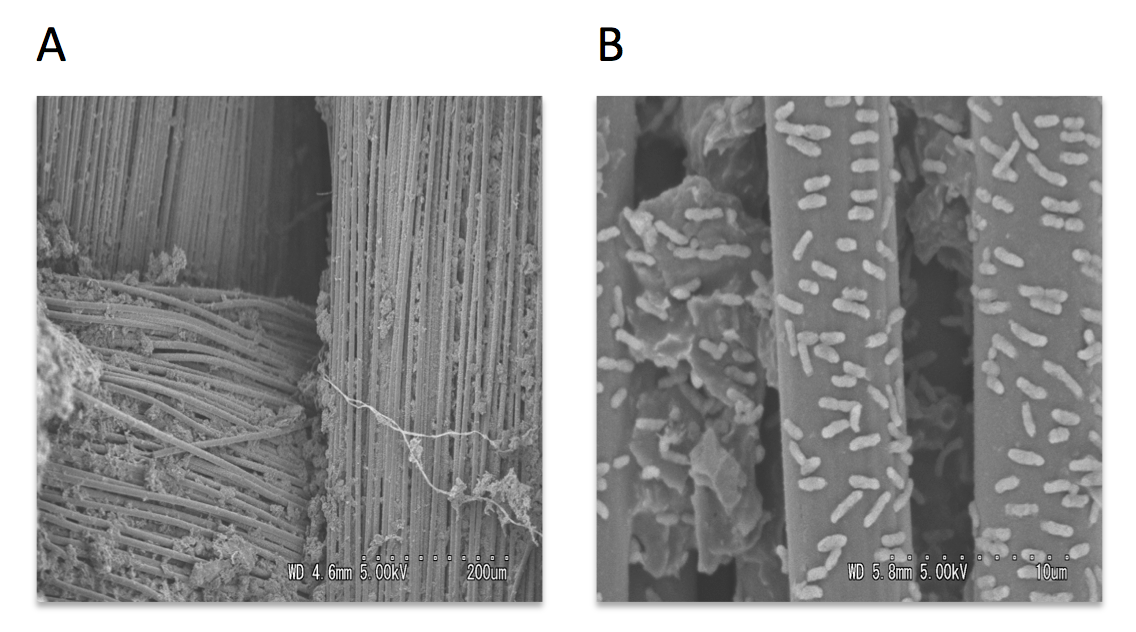
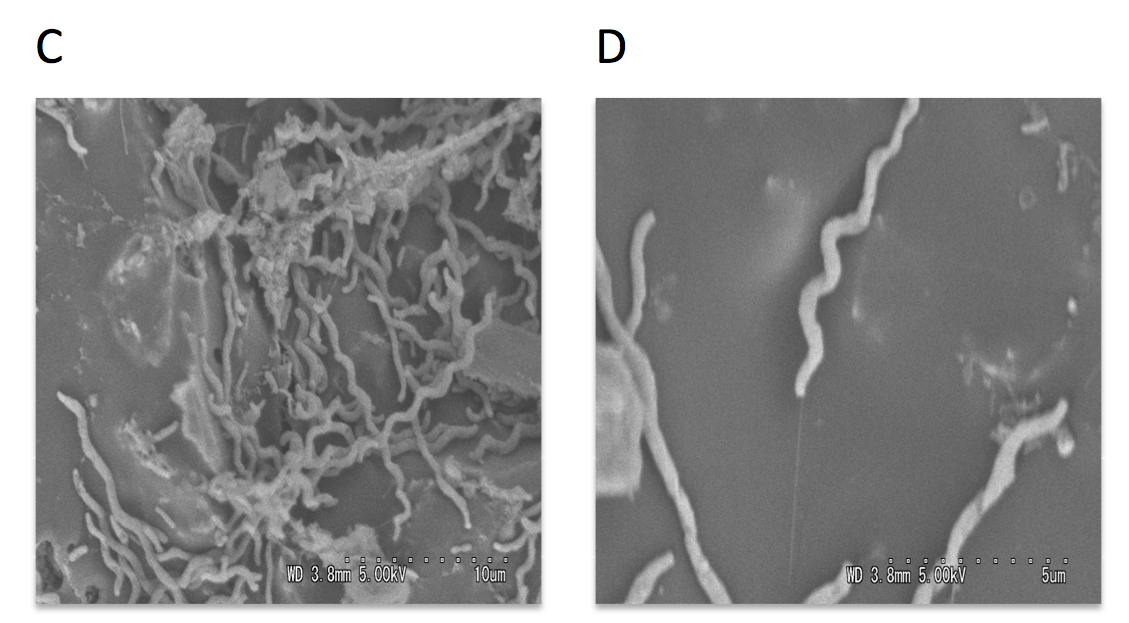
Figure 3: Scanning electron micrographs of the MFC anode surface (A), higher magnification of the anodic carbon fibers colonized with rod-shaped microorganisms (B), anodic carbon granules with attached helical-shaped bacteria (C), and helical bacteria forming pili (D).
Metagenome analysis determined both taxonomic composition and metabolic potential of newly isolated electogenic microbial community. Compared to the reference sample (marine sediment), which was heavily overrepresented by Gammaproteobacteria, the anodophilic microorganisms showed significant increase of Alphaproteobacteria as well as representatives of classes Bacteroidetes and Firmicutes (Fig.4).
A
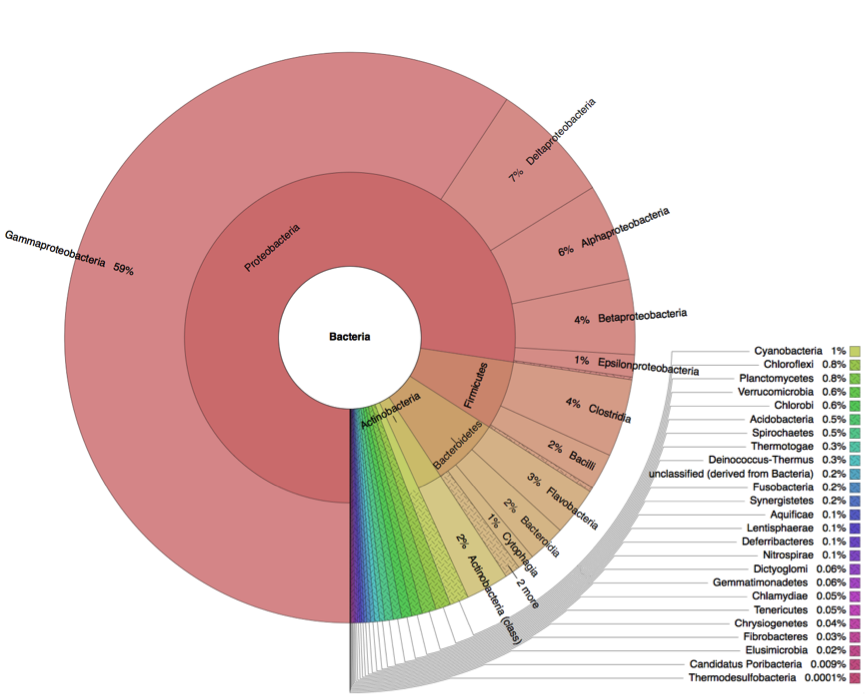
B
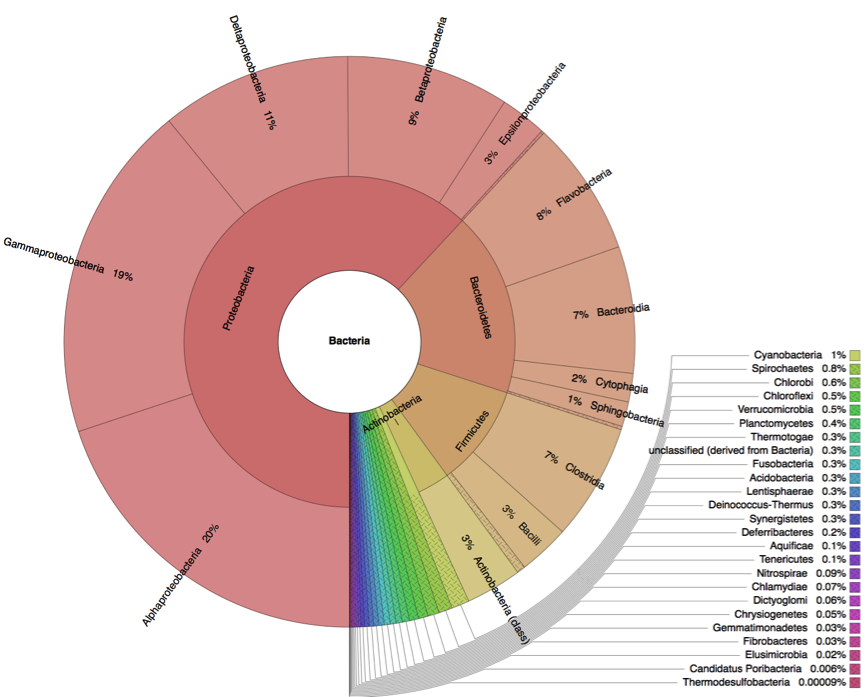
Figure 4: Distribution of bacterial classes in the initial marine sediment (A) and anodic biofilm (B).
Despite of drammatic changes in microbial composition under MFC selective pressure, the anodophilic association did not lose any of the unique biodegradation pathways that are essential for breaking down organic pollutants and toxic compounds by means of microbial metabolic activities (Fig.5).
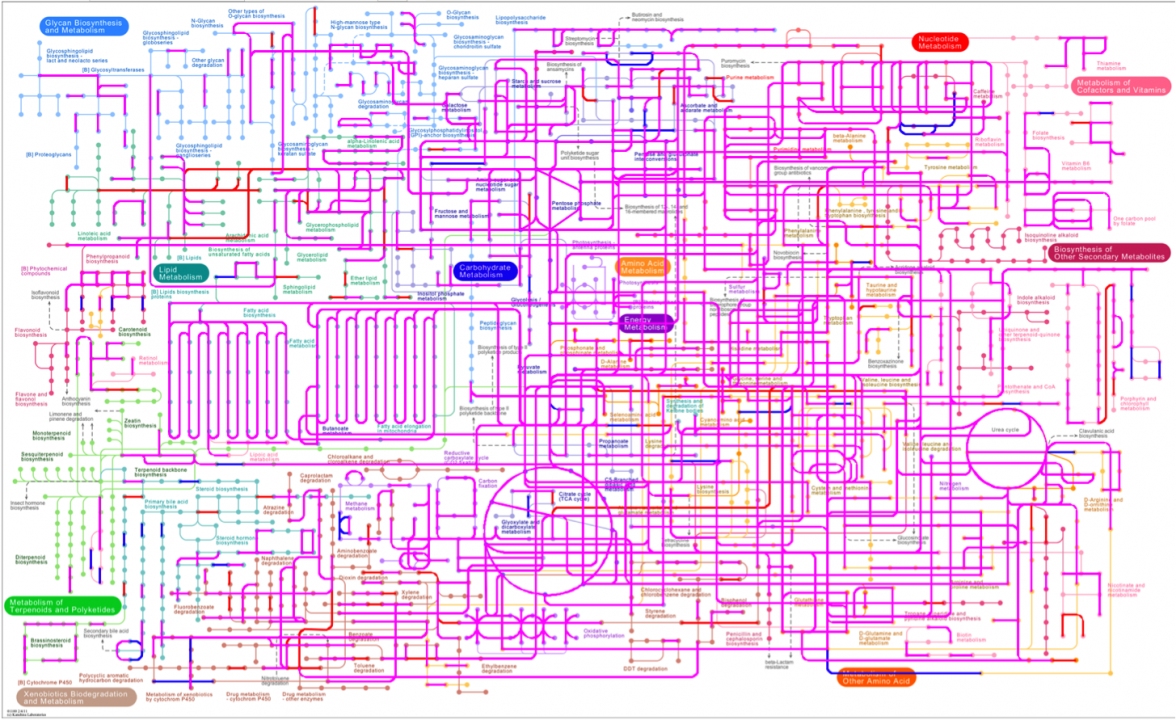
Figure 5: Projection of the global metabolic profiles onto KEGG pathways. Pathways unique for the reference marine sediment microbiome are represented in blue, and pathways unique for the anodic biofilm are represented in red. Metabolic pathways common for both microbial associations are shown in purple.
3.2 Human Metabolic Network reconstruction
Complex organisms, including human, comprise of well differentiated cells and tissues that have have different metabolic objectives and play diverse roles in metabolism. Further, eukaryotic cells contain well differentiated organelles to carry out different physiological functions. Due to localization of metabolic enzymes, many metabolic processes involve coordinated interactions between different organells, and a large number of reactions may occur only in a specific organelle. To gain a complete understanding of the human metabolism, we improved previously reconstructed Edinburgh Human Matabolic Network (EHMN) by adding information on reaction subcellular location and tissue distribution. The resulting database is available at http://csb.oist.jp
3.3 Albatross: dynamical local and global sensitivity library for bioinformatics
Albatross is a Parallel Global/Local Sensitivity Analysis software tool. The software is an open source library written in standard C++ and parallelised with MPI. Several variance-based global sensitivity analysis methods, Sobol and the extended Fourier Amplitude Sensitivity Analysis Test (eFAST) and Partial Ranking Correlation Coefficient, are implemented. Both global sensitivity analysis and local sensitivity analysis are supported, and the package enables a dynamical, time dependent analysis of the sensitivities. The library has ordinary differential equation (ODE) solvers and simultaneous equation solver, the integral and derivative functionality, so that the model output variables can have complicated mathematical representation and many complicated models, including the cascade models, can be analysed.
In recent years, quantitative models requiring numerical simulation have been increasingly applied to various fields, including engineering, economics, biology and medical study. Those models are becoming ever more rich and complicated as they describe complex systems and phenomena with unprecedented detail and accuracy. These models can become difficult to simplify into simpler models describing only the most relevant dynamics of a given problem.
To evaluate and verify a complicated model, it is essential that the response of model results to collective variation of the model input parameters be analyzed quantitatively using sensitivity analysis. The purpose of the sensitivity analysis is to identify the model input parameters that most significantly determine the model output variables. The mathematical methods and computational algorithms of sensitivity analysis have been developed and applied to widely variety field described below.
There are two kind of sensitivity analysis: Local and Global. Local sensitivity analysis measures the response of model output to a small perturbation in the each input parameter’s given value independently; however this does not obtain information from the full search space. The Global sensitivity analysis, in contrast, is able to obtain information of all model input parameters searching spaces even if these parameters are correlated each other.
The models under consideration for local or global sensitivity analysis often have large numbers of input parameters, may have output is not always unique, or may be functions of several dynamical variables. A model can be difficult to simplify and be consequently remain represented in a computationally time-consuming form, such as a large set of simultaneous ordinary differential equations (ODEs).
Further, which model parameters most significantly determine the dynamics and hence the model output, may in fact be time dependent, and our sensitivity analysis generates the full time dependence of this sensitivity. Albatross can analyze any models involving many input parameters even if the model has time-consuming representations. The library obtains a significant performance speed up because it is paralleled with MPI. This enables portability to many contemporary systems, from a general multicore PC to cluster systems and high performance parallel computers.
Albatross supports several global sensitivity analysis methods: the Sobol method extended Fourier amplitude sensitivity analysis test (eFAST) and Partial Ranking Correlation Coefficient (PRCC) are implemented.
The Latin Hypercube Sampling (LHS) for sampling model input parameters with uniform and Gaussian distributions is available as well as sampling with Sobol pseudo-randoms and general random numbers. Albatross has ODE solvers and a static simultaneous equation solver. For any given a model, the both derivative with respect to time (or with respect to any dynamical variable) of the model output, and the single or multiple integrals may be calculated.
General numerical calculations as model functions are provided through GNU scientific library (GSL), FFTW, Sundials libraries and LSODE C++ library. Albatross can make use of several ODE solvers, the sundials ODE solver, CVODE, and LSODE developed by Livermore laboratory. LSODE was originally written in Fortran, but we have translated this to C++ and this is provided in the library the author. This version of the LSODE C++ library is available to download in the same package as Albatross.
User options such as the required precision for a solver, sampling methods, sensitivity analysis method, and the number of simulations, are passed as command line options to the executable, or an input XML file. A model which has a complicated argument structure may be given by user generated code in a suitable form to Albatross. Albatross is also able to accept SBML model desciptions, which have been increasingly popular in various field. For this operation, the SBML and XML2 library are required.
Albatross is a parallel library written in C++ and MPI, so that a standard C++ compiler and MPI library are required to compile and build.
4. Publications
4.1 Journals
- Hao, T., Ma H., Zhao, X., and Goryanin, I. The Reconstruction and Analysis of Tissue Specific Human Metabolic Networks. Mol. BioSyst., 8:663-670, Feb 2012
- Sorokin A, Selkov G, Goryanin I. A user-defined data type for the storage of time series data allowing efficient similarity screening. Eur J Pharm Sci. 2011 Dec 9.
- Lebedeva G, Sorokin A, Faratian D, Mullen P, Goltsov A, Langdon SP, Harrison DJ, Goryanin I. Model-based global sensitivity analysis as applied to identification of anti-cancer drug targets and biomarkers of drug resistance in the ErbB2/3 network. Eur J Pharm Sci. 2011 Nov 9.
- Karelina TA, Ma HW, Goryanin I, Demin OV. EI of the Phosphotransferase System of Escherichia coli: Mathematical Modeling Approach to Analysis of Its Kinetic Properties, Journal of Biophysics, doi:10.1155/2011/579402, Oct 2011
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- Kiseleva L., Goryanin I. Microbial fuel cells as a new source of green energy. The second international workshop "Toward the development of an R&D cluster in Okinawa". March, 2012. (poster)
- Goryanin I. Sustainable Organics Processing - the natural solution. MFC presentation at OIST. March 2012.
- Kiseleva L. Microbial fuel cells - a new source of green energy. Junior research retreat in OIST. December, 2011.
- Kiseleva L., Goryanin I. Computational analysis of metagenomes and metatranscriptomes: challenges and opportunities. ISMB, July 2011. (poster)
- Kiseleva L., Goryanin I. Metagenomic analysis of exoelectrogenic bacteria that power microbial fuel cells. MCCMB, July, 2011. (poster)
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
Nothing to report
7. Other
Nothing to report.



