Deigo Migration Guide
This site is discontinued.
Please go to SCDA-guide.oist.jp
Welcome to Deigo!
First, how to log in:
-
Use "deigo.oist.jp" for logging in, copying files and so on. "sango.oist.jp" is no more.
-
Your user name and password is the same.
-
Your SSH keys will continue to work as ususal.
Each section below deals with a different part of migrating to Deigo. Each one has a summary with the main points, followed a more detailed discussion. We suggest that you at least quickly scan the main points.
Storage
Migrate from Sango Work
Partitions and cores
Running jobs
Software Modules
Building your own software
Storage
Our storage systems have been refreshed and reorganized.
- Bucket is available read-only directly from the compute nodes.
- There is no Work storage. Flash, available as /flash, is a smaller and much faster system for temporary storage during jobs.
- All data must be kept in Bucket. Don't leave any data in Flash.
- You have read-only access to Deigo storage from Saion and vice versa.
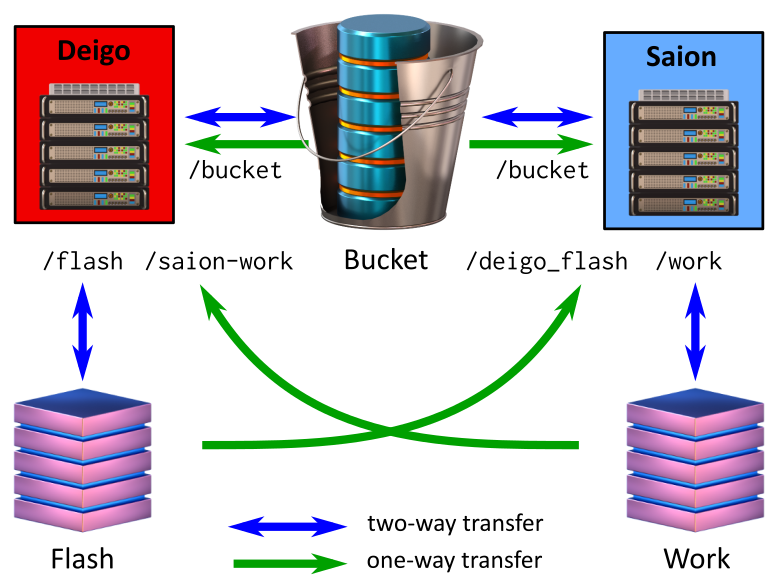
This is Deigo storage:

The Deigo storage organization. Flash is available read-write from both compute nodes and login nodes. Bucket is read and writeable from the login nodes, but read-only from the compute nodes.
Old /work will be available as "/sango-work" for a limited time only. Make sure to go through your folders and copy all the data you need from /sango-work to bucket before it disappears. Focus on the data you need for the next few weeks or so,
The changes to the storage system means that you need to adjust the way you run jobs. The Flash system is too small to store data long term. Store research data permanently on Bucket. You read that data directly from Bucket when you need it, store temporary data on Flash during processing, then move the results back to Bucket and clean up Flash.
Please see the section on running jobs for more details and examples.
Storage access between Saion and Deigo
Flash is available read-only on the Saion nodes as "/flash", and Saions Work is available read-only on Deigo as "/saion-work":
If you are running a pipeline that includes both CPU and GPU steps, such as Relion, you can now keep your original data on Bucket and intermediate data on /flash or /saion-work while using Deigo for CPU computation and Saion for GPU tasks.
With a bit of trickery you can even submit Saion jobs from Deigo (or the other way around). You can use "ssh" to run commands on a remote computer. So if you normally start a job like this on Saion:
$ sbatch myjob.slurmYou can from Deigo do:
$ ssh <your-name>@saion.oist.jp sbatch myjob.slurmMigrate from Sango Work
Sango Work storage will disappear. The storage system is end of life and is too slow for use on Deigo. It is available as /sango_work read-only for a limited time (until the end of the year or so) so you can move or clean up any data remaining on it before it disappears.
First, focus on what you need right now. Clean up Bucket to give you space. Then move any data you need for the next few weeks from Work to Bucket. You want to make sure your work is not interrupted too much by this transition.
Next, take a look at your data:
- The data is "junk". This includes failed simulations and analyses; temporary files, object files and logs; and duplicate data already stored on Bucket. This data can all safely be deleted.
Once Deigo is operational the file system is read-only so you can't erase anything. But you can email us and ask us to delete the data for you.
- Research data you no longer need. Data from completed projects or data left behind by former unit members. If you don't need the data, and you don't want to clutter up Bucket with it you can ask us to archive it.
- For research data you need now or in the future, copy it over to Bucket. Then please ask us to delete the old copy on Work.
Once Work is decommissioned, any left-over data will be permanently archived as a safety measure. However, this is only a legal precaution; you will not be able to request data back from this archive in the future. Please make sure that you have gone through and deleted, archived or copied everything before that happens.
Transfering data from Work
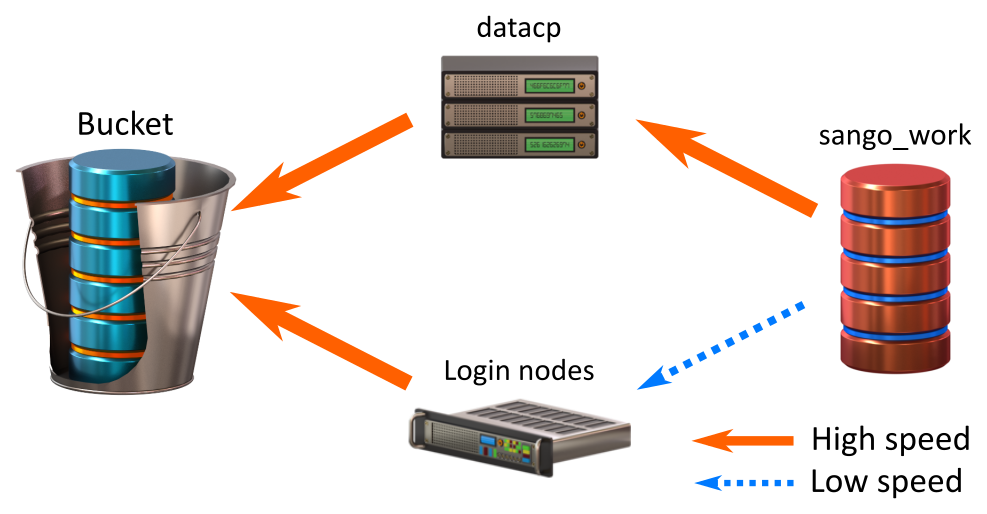
Transfer speed between Bucket and the different Deigo nodes is very fast. Transfer between Sango Work and the datacp partition is also fast. The connection between Sango Work and the Deigo login nodes, however, is quite limited.
Also, all filesystems, including Bucket, deal better with larger, fewer files than with many small files. Copying, storing and searching lots of small files tends to be very time consuming.
If you have a limited amount of data, and a limited amount of files — Say, a Gigabyte or so, and a few hundred files — you can simply copy it all from /sango_work to /bucket using "cp" on the login nodes.
If you have lots of data and many files we strongly recommend that you use the datacp partition, and use tar to make a single archive instead of folders full of files.
Let's say you have a folder full of data in the /sango_work/MyunitU/sim_results/ folder. It's several Gigabytes and thousands of files. You want to copy it to /bucket/MyunitU. Start a job on the datacp partition, and use "tar" to make an archive as you copy:
$ srun -p datacp -t 0-12 --pty bash
....
$ cd /sango_work/MyunitU
$ tar -czf /bucket/MyunitU/sim_results.tgz sim_resultsWe start an interactive job on datacp for 12 hours, and get a command line on it. The default memory and cores (4GB and one core) is plenty for copying data.
We go to where our data is ("/sango_work/MyunitU"). We then run tar, and tell it to create ("c") a compressed ("z") archive in /bucket/MyunitU/sim_results.tgz, and put everything in "sim_results" into it. The "f" option specifies the name of the archive file.
For really large sets of data (many terabytes) we recommend you create a batch job so you don't have to stay logged in as it works. A batch script would look like:
#!/bin/bash
#SBATCH -p datacp
#SBATCH --time 5-0
cd /sango_work/MyunitU
tar -czf /bucket/MyunitU/sim_results1.tgz sim_results1
tar -czf /bucket/MyunitU/sim_results2.tgz sim_results2
tar -czf /bucket/MyunitU/sim_results3.tgz sim_results3
With really large sets of data it's a good idea to create multiple archives with files that belong together. That way you would not have to unpack everything just to look at a small subset.
If you don't immediately need the data, just leave it as an archive. It is much easier and faster to deal with a single archive file than folders full of thousands of files. Also, the archives are compressed so they may save you a fair bit of space.
If you want to unpack a tar archive, you only need to do "tar -xf":
$ tar -xf sim_results.tgz
$ ls
sim_results/ sim_results.tgzWe recommend that you use "tar" to store large folders full of files in general. It's a convenient way to save yourself a lot of time and trouble.
Partitions and Cores
Deigo has two public partitions, "short" and "compute", with different maximum number of cores and time. Deigo also has two different types of nodes, made by Intel and AMD. "short" is for short jobs, interactive jobs and jobs that can use lots of cores. "compute" is for longer multi-node jobs.
Deigo Nodes
Deigo has two kinds of nodes, "AMD" and "Intel". The AMD nodes each have 128 cores and 512GB memory. The Intel nodes have a maximum of 40 cores and 512 GB memory.
The Intel nodes can be faster for specific operations, such as matrix multiplications. On the other hand, the AMD nodes in the "short" partition tend to have faster I/O and have many more cores - 128 instead of 40 - so they are often still faster overall.
The only way to know for certain which system is faster for your particular application is to try running it on both Intel and AMD nodes and compare.
Please see our page on Deigo for more.
Deigo Partitions
Note: This partition layout is premilinary. We will observe how Deigo and the partitions are used in practice, and may change the layout and limits later on if needed.
You now have two partitions available by default: "short" and "compute".
| partition | cores | system | time | notes |
|---|---|---|---|---|
| short | 4000 | All | 2 hours | For short jobs, array jobs and interactive jobs. |
| compute | 2000 | AMD | 4 days | longer multicore jobs. |
The vast majority of jobs on our systems will fit comfortably within these two partitions. You can still request to trade cores for time, as detailed on this page.
In the future there may be other restricted partitions available for jobs that do not fit within the two existing partitions. We want to see how the existing Deigo partitions are used before we decide on any particulars, so there are no details to give you on that at this time.
A note on Memory
The default memory setting is 4GB per core. This is the same default as on Sango. You take more memory the more cores you ask for.
But now Deigo has a lot more cores per node than Sango did - 40 cores on intel, and 128 cores on AMD. As you increase the number of cores you will allocate much more memory than you really need. With 128 cores you would try to get more memory than is available on the node.
Don't rely on the default memory setting. Always explicitly specify how much memory you need, either per core (with "--mem-per-cpu") or per node (with "--mem")!
The "short" partition
The "short" partition covers all nodes on Deigo; including AMD, Intel and Largemem nodes. Which nodes you get depends on where your job will fit. If you ask for more than 40 cores per node, for instance, your job can only be allocated on the AMD nodes.
"short" is a low-priority partition. It overlaps with other partitions on the system, and jobs on "short" can be suspended if a job on another partition needs the cores. If your "short" job runs on a Largemem node, for instance, and a new job on Largemem needs that node, your job will get suspended. This may result in your job failing; if not, it will be resumed once the Largemem job is done.
However, this will only happen if there are no free nodes available for the higher-priority partition. We expect that this will happen only very rarely or not at all. Also, your "short" jobs are guaranteed at least 4 hours uninterrupted running time no matter how busy the systems are.
The Short partition includes the Intel nodes that can use long vector instructions ("AVX512") which can benefit certain mathematical operations, matrix-matrix multiplications in particular. However, programs compiled for the Intel nodes will run only on these nodes, while AMD-built code will run well anywhere. See the section on building apps for more.
You should use the "short" partition if your job needs less than 2 hours, and:
- You are doing interactive work.
- You are running array jobs.
- You run anything that benefits from many cores.
- You want to use AVX512 vector instructions
If you need more time, you can request to trade cores for time, as detailed on this page.
The "compute" partition
The "compute" partition consists of about 45k AMD cores in Deigo, about 5× more than on Sango. A "compute" job can run for up to 4 days, and a user may use up to 2000 cores.
You should use "compute" if:
- You need more than 2 hours.
- You can make use of many cores and multiple nodes in parallel.
For "compute" you can also ask to exchange cores for time. Please see our page on extending running time or cores for more.
Other Partitions
The "largemem", "bigmem" and "datacp" partitions are available as before, and the number of "largemem" nodes have increased. "Largejob" is a new partition with room for very large computations.
Most Largemem nodes have 512GB of memory, the same as those on the "short" and "compute" partitions. But unlike "short" and "compute", there is no running time limit. Also, 14 of the nodes have 768 GB, more than the maximum on "short" and "compute". Largemem is well-suited for long-running bioinformatics jobs that need lots of memory and weeks of computing time.
Likewise, while it is as fast to copy data between the storage systems on the login node as it is to copy data through the "datacp" partition, the "datacp" partition lets you start a data copy job that may take many hours, then log out without having to wait for it to finish.
Largejob has 12800 AMD cores. This partition is managed by the Scientific Computing Committee (the SCC), and you apply for it by asking your unit leader to contact the SCC on your behalf.
Running Jobs
We use Slurm on Deigo, as we did on Sango. For the most part you run jobs the same way you did before. Here a summary of differences:
- Specify "short" or "compute" for your jobs, depending on which one fits you better. You now must specify a partition.
- Be careful with the number of cores and running time. The limits are very different between the two partitions, and the limits you had on Deigo.
- You can read data directly from Bucket, so don't copy it to Flash. Leave your research data in a single place.
- Move your results to bucket and clean up Flash after use. You don't have enough space to store data on Flash permanently.
New Workflow
As the storage layout has changed you need to change your workflow a bit.
First, you no longer need to copy data from Bucket before running a job. Leave your data permanently on Bucket, and read it from there.
Start your job from your home directory. This is the place the "slurm-xxx.out" output files will be saved. Save any data generated during the computation to /flash, just like you used to save data to /work.
At the end of your job, copy the results you want to keep back to Bucket. How do you do that when Bucket is read-only? You can use "scp", like this:
$ scp /flash/MyunitU/mycoolresults.dat deigo:/bucket/MyUni/The syntax is "scp files-to-copy name-of-system:/destination/directory". The colon ":" separates the name of the system and the path to the destination. Without a colon, "scp" will just copy to a local file with that name. The "-r" option ("recursive") to "scp" will copy an entire directory and all its contents.
Finally, clean up all the data you created during your job. /flash is not large, so if you don't clean up it will soon fill up and you and your other unit members will be unable to run anything.
A good way to do this is to create a temporary directory on Flash for this job, do all computations in there, then clean up everything at once by deleting the directory. Here is an example Slurm script:
#!/bin/bash
#SBATCH -p short
#SBATCH -t 0-1
#SBATCH --mem=20G
# create a temporary directory for this job and save the name
tempdir=$(mktemp -d /flash/MyunitU/myprog.XXXXXX)
# enter the temporary directory
cd $tempdir
# Start 'myprog' with input from bucket,
# and output to our temporary directory
myprog /bucket/MyunitU/mydata.dat -o output.dat
# copy our result back to bucket. We need to use "scp"
# to copy the data back as bucket isn't writable directly.
scp output.dat deigo:/bucket/MyunitU/
# Clean up by removing our temporary directory
rm -r $tmpdir Here the first line, "mktemp -d" creates a directory with a guaranteed unique name (the "XXXXX" part is replaced with a unique random string). You can be sure you won't accidentally mix data with another of your jobs. We store the name in "tempdir".
We then cd into this directory, run our job, then use "scp" to copy our results back to bucket. Finally, we remove the entire directory, making sure that we clean up everything properly.
Pipelined computations
A few types of computations involve running multiple applications, one after another, with very large intermediate data files. If these data files are large enough, the 10TB allocation on Flash may not be enough to store all of it at once.
The solution is this case is to move temporary files to Bucket in between the computation steps. We reuse the idea with temporary directories above: Create a second temporary directory on Bucket, use it for temporary storage of these intermediate files, then delete it to clean up at the end. Here's an example script:
#!/bin/bash
...
# create a temporary directory for this job and save the name
tempdir=$(mktemp -d /flash/unitU/myprog.XXXXXX)
# create another temporary directory on Bucket
bucketdir=$(ssh deigo mktemp -d /bucket/MyunitU/myprog.XXXXXX)
# enter the temporary directory on Work
cd $tempdir
# Start 'myprog1' with input from bucket,
# and output to our temporary directory on /flash
myprog1 /bucket/unitU/mydata.dat -o output1.dat
# copy the output file to the bucket temporary,
# then delete it from flash:
scp output1.dat deigo:$bucketdir
rm output1.dat
# run the next pipeline step with output1 as input:
myprog2 $bucketdir/output1.dat -o output2.dat
# repeat as needed
...
# copy final results back, then delete the temporary directories:
scp output.dat deigo:/bucket/unitU/
rm -r $tempdir
ssh deigo rm -r $bucketdir
We can't create or delete a directory on bucket directly as it is read-only from the compute nodes. Instead we use 'ssh deigo <command>' to run <command> on a Deigo login node. We do that when creating a temporary directory, and when deleting it at the end.
OpenMPI
We have built the Deigo software with OpenMPI 4.0.3, a far more modern version than the very old 1.10 version we had on Deigo. We should see a substantial speed improvement and better scaling as a result.
1.10.1 is available as a module, but please avoid using it if you can. It's only there for compatibility with a few very old applications.
We now use the PMIx process manager for OpenMPI:
srun --mpi=pmix <your program>In case your code directly uses pmi2 (this is quite rare) you can still specify it just like you did on Sango:
srun --mpi=pmi2 <your program>
Software Modules
- We have rebuilt and reinstalled all software that was in common use on Sango.
- Modules are now organised into a few different areas.
- We are using a new, different system for modules on Deigo. It's compatible with the old module system but brings a number of improvements.
The module system
We are using a newer version of the module system, and we have reorganised the modules on Deigo. We now have a common area where most software is installed, and four specialised areas.
| Module area | Purpose |
|---|---|
| common | The default area. Most software is installed here. |
| intel-modules | Software that runs best or only on the Intel nodes. |
| amd-modules | Software that runs best or only on the AMD nodes. |
| sango-legacy-modules | The old Sango modules and the "sango" contaienr. |
| user-modules | User-maintained modules. |
To use, say, amd-specific software, you load the "amd-modules" metamodule:
$ module load amd-modulesIf you then look at available modules:
$ module av
------------------------------ /apps/.amd-modulefiles81 --------------------
aocc/2.1.0 aocl.gcc/2.1 gmap-gsnap/2020-04-08 (D)
aocl.aocc/2.1 gmap-gsnap/2014-10-16 gromacs/2020.1
-------------------------------- /apps/.metamodules81 ----------------------
amd-modules (L) intel-modules sango-legacy-modules user-modules
-------------------------------- /apps/.modulefiles81 ----------------------
BUSCO/3.0.2 comsol/51 matlab/R2013a
Gaussian/09RE01 comsol/52 matlab/R2013b
HTSeq/0.9.1 comsol/54 (D) matlab/R2014a
MaterialsStudio/2016 crystalwave/4.9 matlab/R2014b
MrBayes.mpi/3.2.3 csds/2016.0 matlab/R2015a
...The AMD-specific modules are now listed first, followed by the metamodules for the different areas, and then the common modules. You could now load, say, "gromacs" for AMD:
$ module load gromacsYou can load multiple metamodules at the same time. If they have modules of the same name, the module in the last loaded metamodule is picked. If you would load "intel-modules" now, then load "gromacs", you would get the version built for the Intel nodes.
NOTE: modules in "amd-modules" will work on all systems. Modules in "intel-modules" will be fast on the Intel nodes, but will crash on AMD nodes and Largemem. The exception is the intel compiler, which works everywhere.
Make sure that you use the appropriate version of your software when running your jobs - if you are using the "short" partition it's probably best to stick with common modules and those in "amd-modules".
The new module system, called "lmod". has a lot of improvements. You can read about it on our updated module page, but here is a short overview:
There is a new short-form command called "ml". It's very convenient. The "module" command is still here and works the same as always. What are the differences between "module" and "ml"?
"module" is designed to work exaxcly like the old module command. It is great to use in scripts, as you spell everything out so it is completely clear what you are doing.
"ml" is a separate command, and is very efficient for interactive use.
Just "ml" will list your loaded modules. "ml python" will load python. "ml -python R" will unload python (note the minus sign), then load R. But "ml" also takes all commands that "module" does. You can say "ml load python", or "ml list" if you want.
Here's a summary, with the equivalent "module" command and the effect:
| command | full module command | effect |
|---|---|---|
| ml | module list | lists modules you have loaded |
| ml |
module load |
loads |
| ml - |
module unload |
unloads module (note the minus sign) |
| ml av | module available | lists available modules |
| ml
|
module
|
you can use any 'module' command as usual |
You can list several modules to load and unload. They are loaded in the order you list them. To, say, unload amd-modules, and load the intel node version of GROMACS, you could simply do:
$ ml -amd-modules intel-modules gromacsThis unloads amd-modules, loads intel-modules, then finally loads gromacs in the intel area.
You can "save" and "restore" lists of modules that you often use together. Instead of adding modules commands to your ".bashrc" startup script, save the modules you usually need, and restore them when you need them.
If I often use "python/3.7.3" and the "amd-modules" metamodules, for instance, I load them once, then use the "save" (or "s") command to save them to my "default" list:
$ ml python/3.7.3 amd-modules
$ ml saveWhenever I want to load them, I can do ml restore or ml r to reload them. You can give lists a name, and have multiple lists for different tasks.
We're trying to add helpful information to the modules. You can do ml help <module> to get a hopefully informative help text about many modules. If you feel there's something missing or wrong, please let us know and we'll fix it.
We're also adding keywords to each module to make them easier to find. You do ml key genomics to find genomics-tagged modules for instance. Again, if you think a module is missing a keyword, or is miscategorized, let us know!
Deigo Software Modules
We have rebuilt all commonly used software on Sango for Deigo. In addition we have also installed the newest version were applicable.
-
The paths to the software has changed, and the versions may not be exactly the same as before. You will need to edit your slurm scripts accordingly.
-
Most software is installed in the common area. However, a few modules that benefit from specific hardware is installed in amd-modules and intel-modules. If you do not see your module in the common area, look in the amd and intel-specific areas as well.
-
The software has been rebuilt, but we can't exhaustively test each module. If you find a faulty module, please let us know as soon as possible.
-
We have usually installed the latest Sango version on Deigo. If you used to use an older version, please try the newer available version to see if it fits your needs.
-
Older versions of software such as R may no longer have all packages available online, and will miss functionality that was present on Sango. A few software modules can't be built on Deigo or are no longer available for download at all.
Python
Python 2 is officially end of life. We strongly recommend that you use Python 3 whenever possible. You should only use Python 2 for running old, unmaintained software that was never ported to version 3. The installed version is 2.7.18, the last and final release. We will not maintain or update it further.
jupyter is now installed as part of the python and R installations. The "jupyter" module just loads the latest python and R, and gives you a short instruction on how to start. You can use jupyter without the module by loading python, then running "jupyter-notebook".
When you run python commands, make a habit of always using the version number. So "python3"; "pip3"; "ipython3" and so on, not just "python" or "pip". This ensures that you really run the version of python that you intend.
On Deigo there is no plain "python" command. You always need to specify "python3" (or "python2" if necessary). For some python software, you may need to edit the source slightly to reflect this:
If you try to run a python application and it complains that "python" can't be found, you need to open the program file with an editor. The first line says something like:
#!/usr/bin/env python
Change "python" to "python2" or "python3" as needed. Please contact us if you would like assistance with this; we're happy to help.
Sango software
You can still run original Sango modules as well as your own Sango software directly or using a "sango" container.
Sango Modules
The original Sango modules are available under the "sango-legacy-modules" metamodule. These are available best effort only. We do not maintain these at all. Some of them definitely do not work on Deigo. Many of them will be slow or may exhibit odd problems.
We strongly recommend using software built for Deigo if available.
If you do want or need to use a Sango module, first load the metamodule, then load your software as usual:
$ ml sango-legacy-modules
$ ml qiime/1.9Your own Sango software
Software that you installed on your own may work as-is under Deigo. Your home directory is available as before, and software you installed there will be fairly likely to work without change. However, we recommend that you rebuild it for Deigo if possible.
/apps/unit
Your old /apps/unit/MyUnit directories are now available read-only from the /sango_apps directory on the login nodes only. You can't run software from there (it's not available on the computer nodes), and you have only a limited time to move software and data from there to the new system.
Your unit should already have a new, empty /apps/unit/ directory. You will need to move your software from the old directory yourself.
- The preferred way is by reinstalling the software into the new /apps/unit directory. See below for more dtails.
- You can also copy it straight over, and it may work with some limitations.
/apps/unit now has a size limit, and the maximum size per unit is 50GB. This is plenty of space for software — the entire Deigo software repository uses less — but it's not enough for installing large data sets that some software needs. We suggest that you install this data or the entire software package into /bucket instead.
Using old Sango Software
We recommend that you rebuild and reinstall your software if you can. The modern compilers and libraries that we have on Deigo will give you the best performance and future compatibility. See the section below for tips on building on Deigo.
-
If your version is too old to build on Deigo, consider downloading and using the newest version. A new version is much more likely to work well on a modern system and will have various bugfixes and improvements.
-
If you can't rebuild at all, try running it as usual. In many cases the software will continue to work, but may not be as fast as it could be, and you may run into occasional problems. Software using MPI are especially likely to have issues.
-
If you can't rebuild and it doesn't run, try running it with the "sango" container in the next section.
The "sango" container
We have made a container with the Sango environment to make running old Sango software easier. The container includes the entire original Sango system and libraries, and should make most Sango software run under Deigo. You can use it to run your own software as well as run the old Sango modules.
To use the Sango container you need to load the sango legacy metamodule, then load the "sango" module:
$ ml sango-legacy-modules
$ ml sangoRunning software using the container is a little different than running it directly. You let "sango" run it, rather than run it directly. If you have an application "myprog" and you want to run it with the sango container, run it as:
sango myprog
You can't load modules yourself. Instead you need to tell the container to load modules for you. Let's say your program "myprog" needs the python and gsl modules. On Sango, your slurm script first loads the modules, then runs "myprog":
module load python/3.5.2 gsl/2.0
myprog mydata.datChange it so you load the "sango" module, then use the "-m" parameter (for "module") to the "sango" command to tell it what modules to load for you:
ml sango-legacy-modules sango
sango -m python/3.5.2 -m gsl/2.0 myprog -o outfile mydata.datThe "sango" module is not a replacement for rebuilding your software. It is not reliable, it will not always work (software using MPI doesn't work at this time) and it will be slower. This is a temporary solution to let you continue your work while you rebuild the software for Deigo.
Build Software on Deigo
We use CentOS 8 on Deigo. This is an up to date version of Linux, with a full complement of libraries installed by default.
On Sango we added a number of modules over the years to offer newer versions of libraries and tools that were missing in the system. These modules are no longer necessary. If you have used such library modules to build software on Sango, you most likely don't need them on Deigo. This includes (and is not limited to):
- cmake
- zlib
- opencv
- gsl
- netcdf
- eigenIf you have been using these as modules, try to rebuild your software without loading the module.
A few libraries and tools are installed as modules. This includes the MPI libraries and numerical libraries (see below) as well as Boost and hdf5. These are installed to take maximum advantage of our specific hardware or to work with the GCC 9.1 compiler.
choose your node type
To build software that will work on any compute node on Deigo, you should build on an AMD node. The login nodes are Intel nodes — if you build software on the login nodes it may not work on the AMD nodes.
The Intel nodes, login nodes included, have a couple of hardware features (called "AVX512") that can speed up some types of math operations. But software built to use those features will only work on the Deigo Intel nodes and nowhere else. When you build on an AMD node you are sure to get an application that will work everywhere.
You should make it a habit to always start an interactive job on a compute node when you build software. That way you are sure you are building for the system you intend to run it on. Here is an example:
$ srun -t 0-2 -p short -C zen2 -c 16 --mem=8G --pty bashThis command will start a 4-hour job on the "short" partition, with 16 cores and 8GB memory. The "-C zen2" option tells Slurm to select a node with tag "zen2" — this are the AMD nodes. If you want to use the Intel nodes, use "-C cascade" instead.
Compilers and Numerical libraries
GCC version 8.3 is the system default compiler on Deigo. However, we also make GCC version 9.1 available as a module. It has several performance improvements, and we recommend using it if you can. All Deigo modules have been built using this version. We will make newer versions available over time.
As an alternative, we also have the Intel compiler (in the "intel-modules" area) and the AMD AOCC compiler (in "amd-modules"). Both can generate code for either Intel or AMD systems.
In our recent experience the Intel Fortran compiler still generates better code than GCC, while for C and C++ the better compiler depends on the code you're building. The AMD AOCC compiler generates good numerical code (for AMD systems especially) but still lags somewhat compared to the other two in general.
If you use the Intel compiler, please note that only Intel/2019_update5 and Intel/2020_update1 actually work on Deigo. The older Intel compilers depend on system library versions that are no longer available on CentOS 8. Also, the 2020 version of MKL (see below) will currently be slow on the AMD nodes.
We have three numerical libraries available: OpenBLAS, AOCL and MKL.
OpenBLAS
OpenBLAS is a modern, high-performance implementation of BLAS and LAPACK. Version 0.3.3 is installed system-wide, and 0.3.9 is available as a module. The module is quite a lot faster. OpenBLAS is sometimes the fastest numerical library for low-level operations, but some higher-level operations can be slow.
If you're unsure what to use, and you don't want to spend a lot of time on this, pick the Open BLAS module.
AOCL
AOCL is AMDs optimized numerical libraries (they work fine on both Intel and AMD CPUs). They are split into two modules: "aocl.gcc" and "aocl.aocc", for the GCC compilar and AMDs AOCC compiler respectively.
AOCL contains quite a few libraries, including Blis (for BLAS); libFlame (for LAPACK); Scalapack; a high-performance replacement to libm; an AMD optimized version of FFTW and so on. Please refer to the AMD AOCL web page for details on the libraries.
Blis has single-threaded and multi-threaded implementations. To use the single-threaded version, link with "libblis" or "blis", and link with "libblis-mt" or "bli-mt" for the multithreaded version. We recommend the multithreaded version for best performance. For more information, see the Blis multithreading documentation.
Blis can be very high performance; on AMD it is the fastest library for large matrix multiplications. The higher-level libFlame is not as far along yet, and many high-level operations such as singular-value decomposition are still quite slow and unoptimized.
Consider using AOCL if you are mostly doing low-level matrix operations, especially if you are using the AOCC compiler.
MKL
The Intel MKL library is a popular numerical library for x86 computers. It is not open source - you need a license to use it, so if you depend on this for your own software you may be unable to use it when you leave OIST.
For a number of years MKL was the fastest numerical library available on X86. Today OpenBLAS and Blis can be as fast or faster for lower-level operations. While not always the absolute fastest, MKL is consistently good especially for high-level operations, making it a good choice for applications that do a very wide range of numerical computing tasks. We build and link Python on Deigo with MKL for this reason.
Note that only versions 2019_update5 and 2020_update1 will work on Deigo.
MKL and AMD CPUs
The Intel MKL library has always been very slow on non-Intel cpus. The library checks the CPU maker and if it's not Intel it deliberately chooses the slowest, unoptimized version of its math functions.
We can get around this by explitly telling the library which type of CPU it should assume that we are using. If we set the environment variable MKL_DEBUG_CPU_TYPE to "5" the library will run very well on our AMD cpus. But you need to leave this unset for our Intel cpus. This will work with MKL version 2019, but no (as of this writing) with the 2020 version.
You do not need to worry about this. Don't set this yourself. We set and unset this environment variable as needed on the different compute nodes, giving you the best speed everywhere. As long as you use the 2019 version of MKL you will get good performance. Note that you can build your code with the 2020 compiler, and then load the 2019_version5 module when running it to get the good performance.
You should use the Intel MKL library if you are using a lot of high-level functions or if you are using the Intel compiler suite; and if you are not worried about being able to use your code without having an Intel software license.
MPI Applications
Our current version of OpenMPI is 4.0.3. By comparison the newest version on Sango was 1.10.1. That is ancient. There have been several years of speedups and bug fixes built into the new version. In informal testing we have seen speed-ups of up to 3x and much better scaling from using a new MPI library version.
We aim to have a somewhat-working OpenMPI 1.10 module available on Deigo. However, 1.10 is so old that it doesn't know about or support our current high-speed network. Instead the library will fall back on safe but slow network connections.
- Your MPI program may fail to work with the 1.10.1 library, due to timing issues.
- Your MPI program may work, but will be quite slow and not scale well.
- Some MPI applications that depend on precise communication timing will likely fail completely.
For this reason we strongly recommend that you rebuild your application using the newest available OpenMPI version on the cluster.
For many applications it should be as simple as loading the new 4.0.3 version of OpenMPI instead of 1.10.1, then do a clean rebuild.
Missing symbols
In rare cases you may get an error about missing or deprecated symbols. This may happen when you are rebuilding a very old application that was never updated for more recent MPI standards.
-
If you can, we suggest you find a newer version of your application and use that. Something so old that they still use the MPI 1 symbols will likely have other issues as well, so a newer version will give you a better experience.
-
If you can't update or if there is no new version available, you need to edit the source code to fix these issues. It is not actually very difficult - you mostly just need to replace the old names with the new. You can find a detailed guide here.
Finally...
Deigo is a large, complex system. It is currently very much a work in progress, and will remain so for a long time after release. We are aware of many issues with the system, and there are undoubtedly many more still unknown to us. We apologize in advance for the trouble this will cause you over the coming weeks and months.
To help us make Deigo the best system it can be, we ask a few things of you:
- If you find an issue, try to document it - what happens, how can you reproduce it, when does it happen, when does it not happen - and let us know. The more we know the more we can fix.
- Please be patient. There is no shortage of things to work on, and it may take some time before we can take care of issues that bother you.
- We will make changes to the system, likely several times, over the next few months. Change is disruptive and frustrating. But will be necessary to improve the system over the long term. For this we can but ask for your forbearance.
- Finally, if you have an idea then tell us! It may or may not actually be a good idea, but at least we'll know about it.
Welcome to Deigo! We hope you will enjoy it here.
/SCDA



