FY2019 Annual Report
Cell Signal Unit
Professor Tadashi Yamamoto

Abstract
The Cell Signal Unit studies molecular and cellular events that are relevant to and important for maintaining healthy life in a variety of environmental conditions. Through the studies, Unit explores the cause of various diseases that include cancer, neuronal disorder, immunological diseases, diabetes/obesity, and defects in development at the molecular level. The Unit characterizes regulation of gene expression, particularly post-transcriptional regulation that includes regulation by microRNA, and long non-coding RNA and RNA binding proteins. The Unit also characterizes protein kinase-mediated cell signaling especially in the control of the brain function such as emotions, learning and memory.
1. Staff
- Dr. Patrick Stoney, Staff Scientist
- Dr. Ken Matsuura, Staff Scientist
- Dr. Akiko Yanagiya, Staff Scientist
- Dr. Kristopher Paraiso Montrose, Postdoctoral Scholar
- Dr. Shou Soeda, Postdoctoral Scholar
- Dr. William Ashworth, Postdoctoral Scholar
- Dr. Olga Elisseeva, Visiting Researcher
- Ms. Saori Nishijima, Technical Staff
- Ms. Risa Ishida, Technical Staff
- Ms. Melissa Germany, Technical Staff
- Ms. Nao Ohmine, Technical Staff
- Ms. Atsuko Sato, Technical Staff
- Ms. Sandrine Anne Laure Burriel, Graduate Student
- Mr. Haytham Mohamed Aly Mohamed, Graduate Student
- Mr. Shohei Takaoka, Graduate Student
- Mr. Hemanta Sarmah, Graduate Student
- Ms. Dina Mostafa, Graduate Student
- Mr. Mohieldin Magdy Mahmoud Youssef, Graduate Student
- Mr. Hong Huat Hoh, Graduate Student
- Ms. Aisulu Maipas, Graduate Student
- Ms. Yuki Nakagawa, Research Unit Administrator
2. Collaborations
2.1 Physiology and Molecular Cellular Biology of the CCR4-NOT complex
- Description: Analyze the physiological and molecular biological roles of each component of the CCR4-NOT complex using gene-modified mice.
- Type of collaboration: Joint research
- Researchers:
- Dr. Keiji Kuba, Department of Physiology, Graduate School of Medicine, Akita University
- Dr. Toru Suzuki, Laboratory of Immunogenetics, Riken Center for Integrative Medical Sciences
- Dr. Toshinobu Fujiwara, Laboratory of Biochemistry, Faculty of Pharmacy, Kinki University
- Dr. Masahiro Morita, Department of Molecular Medicine, UT Health
- Dr. Nahum Sonenberg, Department of Biochemistry, McGill University
2.2 Basic cancer research for prevention and treatment
- Description: Search for substances that contribute to the prevention and treatment of cancer from biological resources, and elucidate its mechanism of action
- Type of collaboration: Joint research I and II
- Researchers:
- I. Representative director Kuniaki Nerome, Bioresource Laboratory LLC
- II. Professor Shinya Ikematsu, National Institute of Technology, Okinawa College
2.3 Mechanisms and physiological roles of CCR4-NOT complex-regulated gene expression in the brain
- Description: Electrophysiological studies of gene modified mice with abnormal social behavior
- Type of collaboration: Joint research
- Researchers:
- Team leader Masaru Tamura, RIKEN
2.4 Study of Cell-to-cell Interaction within Tumor Microenvironment Using An In-Vitro Three Dimensional Pancreatic Cancer Model
- Description:
1. Establishing an in vitro 3D organoid culture for pancreatic ductal adenocarcinoma to study cell-cell interaction within tumor microenvironment
2. Investigating the origin of cancer stem cells in pancreatic ductal adenocarcinoma by live cell imaging using tumor organoid
3. Characterizing the effects of the interaction between stromal cells and cancer stem cells
4. Identifying the targets for tumor-stroma crosstalk and cancer stem cells
- Type of collaboration: Joint research
- Researchers:
- I. Representative director Kuniaki Nerome, Bioresource Laboratory LLC
- Professor Masafumi Nakamura, Assistant Professor Kenoki Ohuchida, Dr. Kazuhiro Koikawa, Department of Surgery and Oncology, Graduate School of Medical Sciences, Kyushu University
2.5 Human immune responses and their modulation in cancer and autoimmunity
- Description: Human immune responses and their modulation in cancer and autoimmunity
- Type of collaboration: Joint research
- Researchers:
- Research Scientist Olga Elisseeva, Laboratory of Immunogenetics, Riken Center for Integrative Medical Sciences
3. Activities and Findings
3.1 Hepatic posttranscriptional network comprised of CCR4-NOT deadenylase and FGF21 maintains systemic metabolic homeostasis.
Whole-body metabolic homeostasis is tightly controlled by hormone- like factors with systemic or paracrine effects that are derived from nonendocrine organs, including adipose tissue (adipokines) and liver (hepatokines). Fibroblast growth factor 21 (FGF21) is a hormone-like protein, which is emerging as a major regulator of whole-body metabolism and has therapeutic potential for treating metabolic syndrome. However, the mechanisms that control FGF21 levels are not fully understood. Herein, we demonstrate that FGF21 production in the liver is regulated via a posttranscriptional network consisting of the CCR4–NOT deadenylase complex and RNA-binding protein tristetraprolin (TTP). In response to nutrient uptake, CCR4–iNOT cooperates with TTP to degrade AU-rich mRNAs that encode pivotal metabolic regulators, including FGF21. Disruption of CCR4– NOT activity in the liver, by deletion of the catalytic subunit CNOT6L, increases serum FGF21 levels, which ameliorates diet-induced meta- bolic disorders and enhances energy expenditure without disrupting bone homeostasis. Taken together, our study describes a hepatic CCR4–NOT/FGF21 axis as a hitherto unrecognized systemic regula- tor of metabolism and suggests that hepatic CCR4–NOT may serve as a target for devising therapeutic strategies in metabolic syndrome and related morbidities.
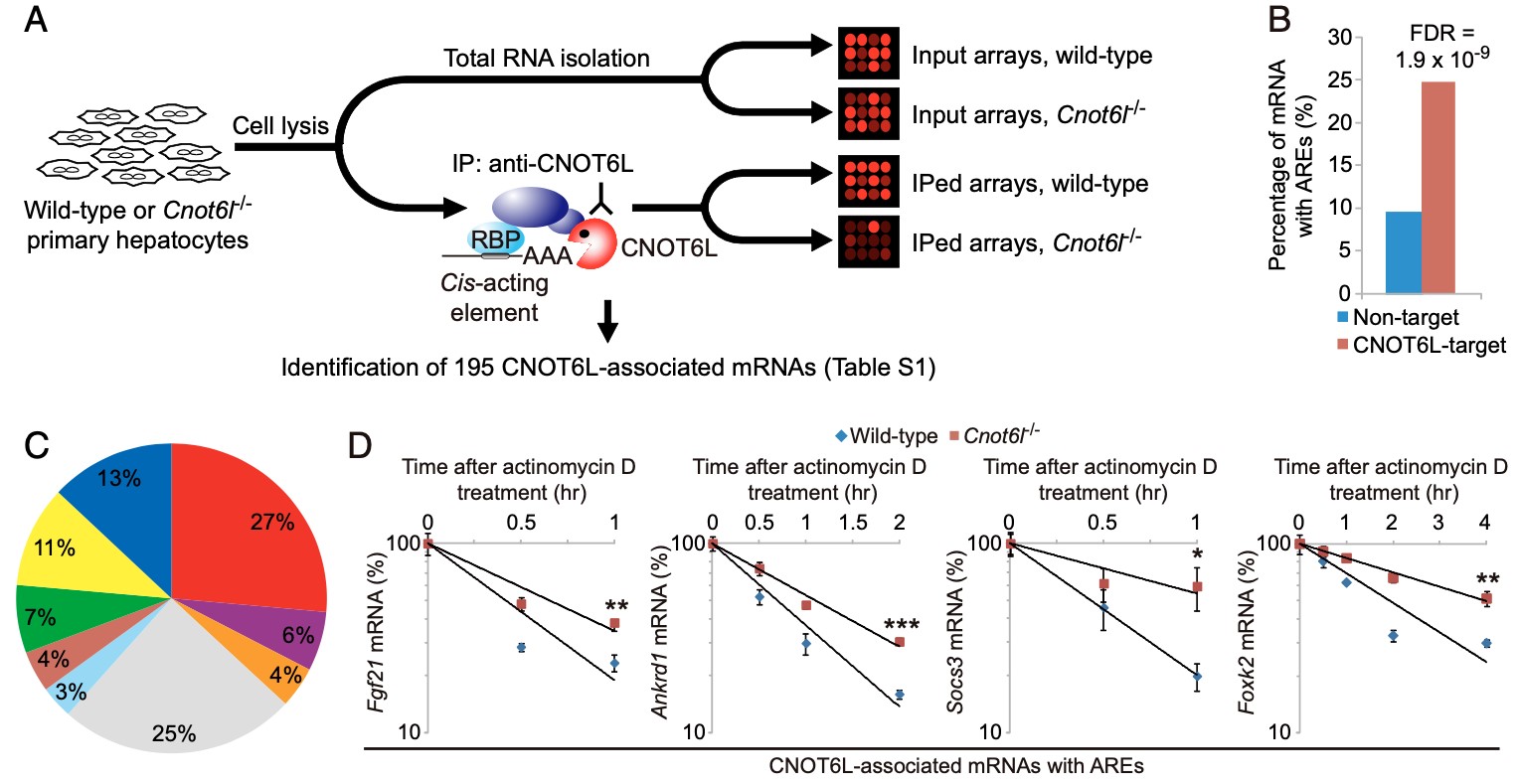
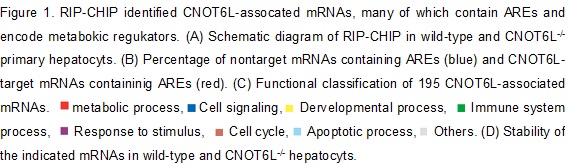
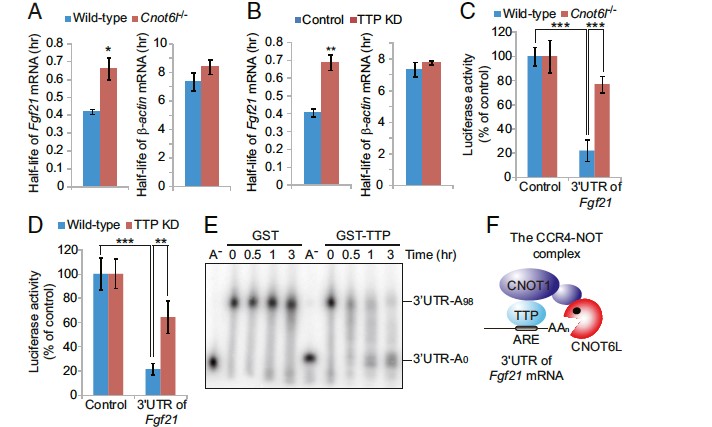
Figure 2. (A) The CCR4-NOT complex degrades the ARE mRNA for hepatokine FGF21 in response to feedings. (B) Half lives, (C & D) Reporter assay. (E) in vitro deadenylation assy in the Krebs Ascites extract. (F) Model for structural organization of Fgf21 mRNA-bound TTP in the complex with CNOT6L deadenylase.
Figure 3. Protection from diet-induced metabolic disorders and enhanced energy expenditure in CNOT6L-/- mice. (A) Growth curves on high-fat diet. Levels of (B) blood glucose, (C) serum triglyserol, (D) serum insulin, (E) serum b-hydrooxybutyrates in mice on high fat diet or fasting. Intraperitoneal glucose torelance tests (F) and insulin torerance tests (G) of mice on high-fat or standard tests. (H) oxygen consumption.
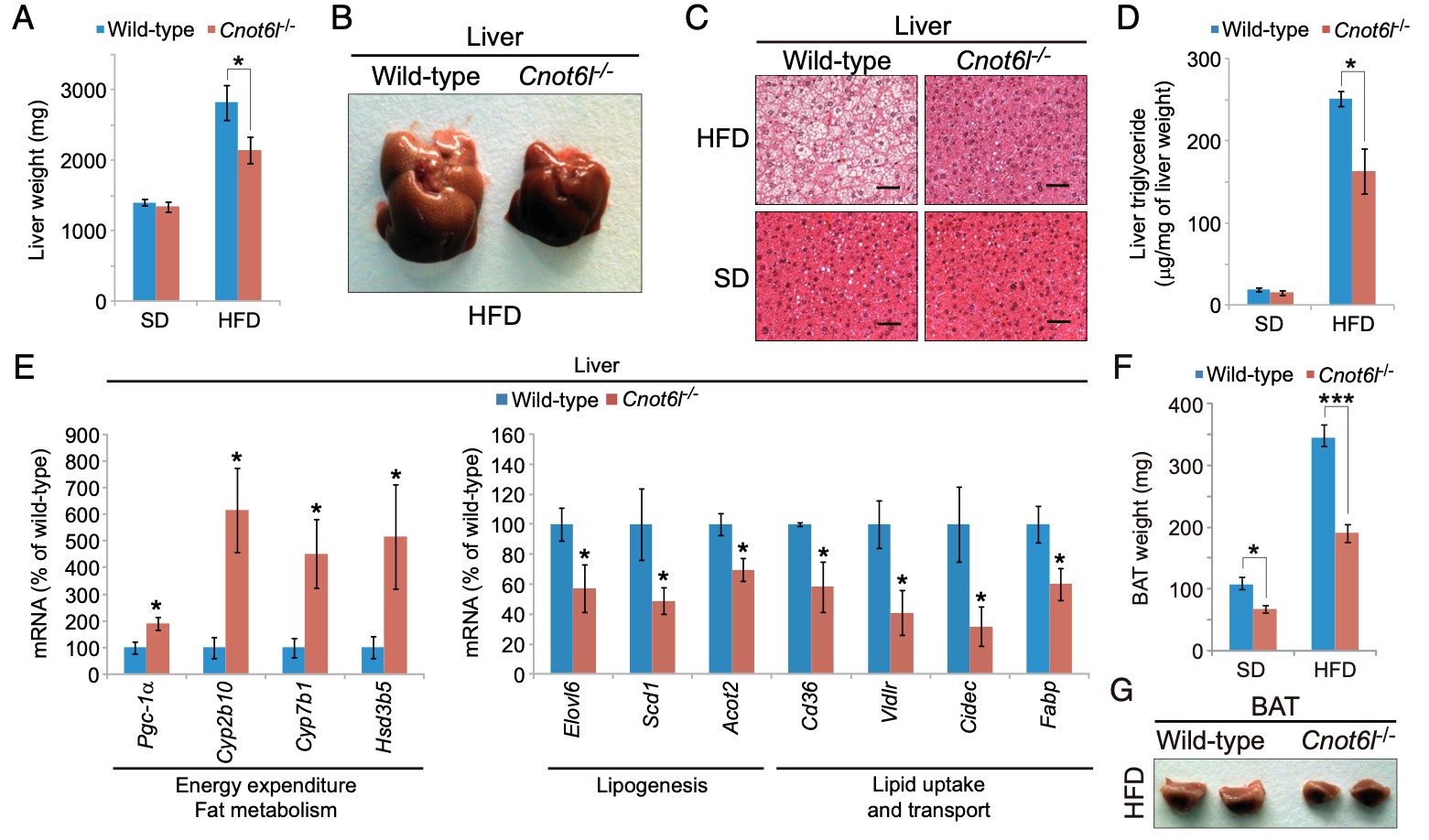
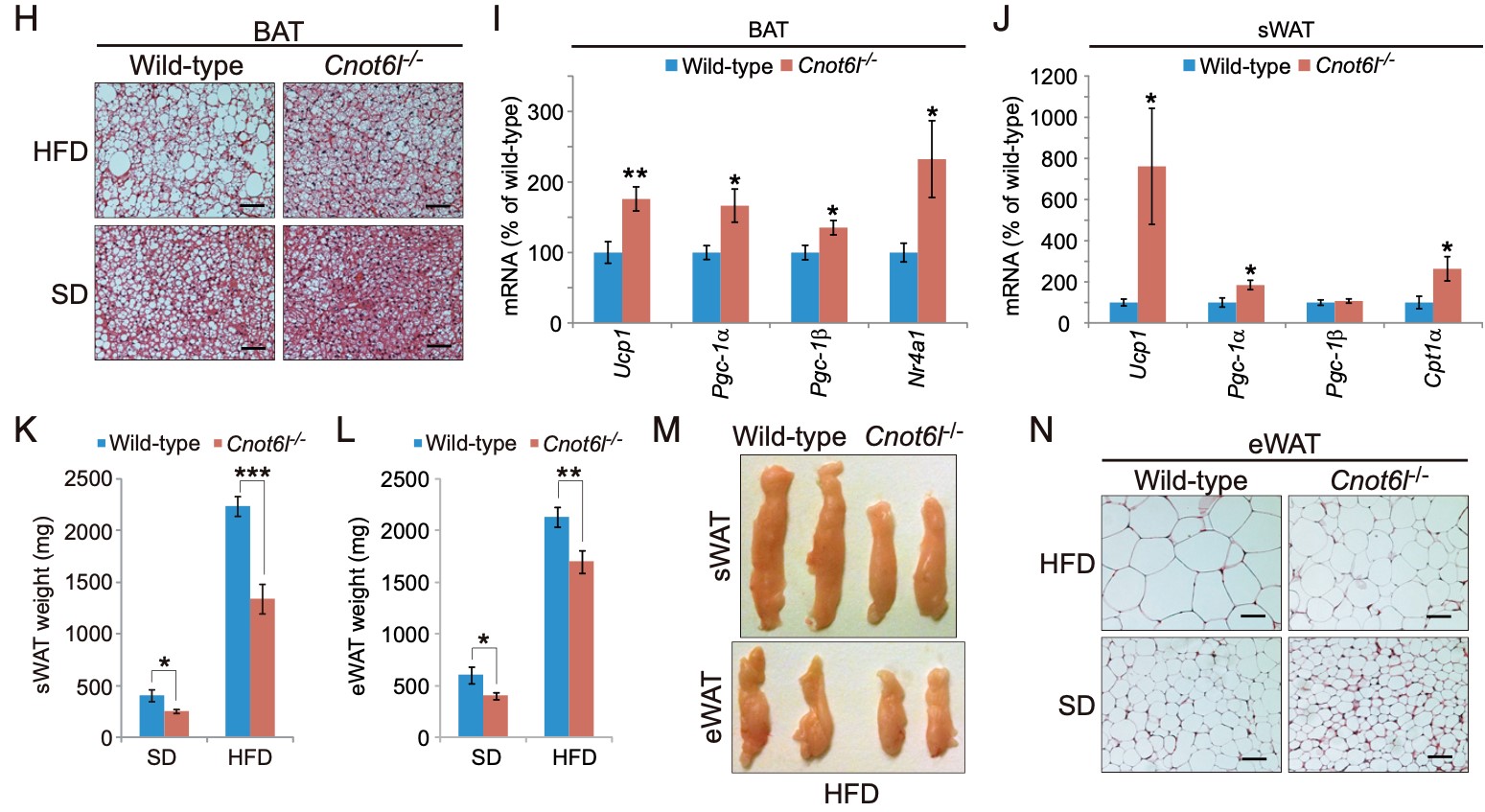
Figure 4. Cnot6l deletion leads to a resistance to lipid accumulation and an increase in energy expenditure in the liver, BAT, and WAT. (A) Weight, (B) representative image, (C) representative H&E staining, and (D) triglyceride contents of livers from wild-type and Cnot6l-/- mice fed on a standard diet (SD) or high fat diet (HFD). (Scale bars, 50 mm.) Liver triglyceride contents were normalized to liver weight. (E) Expression of genes involved in energy expenditure, fatty acid oxidation, lipogenesis, and lipid uptake and transport in the liver of wild-type and Cnot6l-/- mice fed on an HFD. Weight (F), representative image (G), representative H&E staining of BAT (H), and expression of the indicated mRNAs in BAT (I) and sWAT (J) of wild-type and Cnot6l-/- mice fed on an HFD. Weight of sWAT (K) and eWAT (L), representative sWAT and eWAT (M), and representative H&E staining of eWAT (N) from wild-type and Cnot6l-/- mice fed on SD or HFD. Scale bars in H and N, 50 mm.
3.2 CNOT3 targets negative cell cycle regulators in non-small cell lung cancer development.
Lung cancer is one of the major causes of cancer death and clarification of its molecular pathology is highly prioritized. The physiological importance of mRNA degradation through the CCR4-NOT deadenylase has recently been highlighted. For example, mutation in CNOT3, a gene coding for CNOT3 subunit of the CCR4-NOT complex, is found to be associated with T-cell acute lymphoblastic leukemia, T-ALL, though its contribution to other cancers has not been reported. Here, we provide evidence suggesting that CNOT3 is required for the growth of non-small cell lung cancer. Depletion of CNOT3 suppresses proliferation of A549 human non-small cell lung cancer cells with enhanced mRNA stability and subsequent elevated expression of p21. In addition, we identified the mRNA for Krüppel-like factor 2 transcription factor, an inducer of p21, as a novel mRNA degradation target of CNOT3 in non-small cell lung cancer cells. Aberrant up-regulation of Krüppel-like factor 2 by CNOT3 depletion leads to impairment in the proliferation of A549 cells. Consistent with these findings, elevated mRNA expression of CNOT3 in non-small cell lung cancer in comparison with the paired normal lung epithelium was confirmed through scrutinization of the RNA-sequencing datasets from The Cancer Genome Atlas. Moreover, we found an inverse correlation between CNOT3 and CDKN1A (encoding p21) mRNA expression using the combined datasets of normal lung epithelium and non-small cell lung cancer. Thus, we propose that the up-regulation of CNOT3 facilitates the development of non-small cell lung cancer through down-regulation of Krüppel-like factor 2 and p21, contrary to tumor suppressive functions of CNOT3 in T-ALL.

Figure 1. CNOT3 mRNA expression is up-regulated i non-small cell lung cancer (NSCLC). Expression of CNOT3 using the RNA-seq data from TCGA for lung adenocarcinoma (LADC) (a) and lung squamous cell carcinoma (LSqCC) (b). The red bar indicates NSCLC sample and blue bar indicates normal epithelium sample in the same patient (paired normal). RPKM reads per kilobase of exon model per million mapped reads.
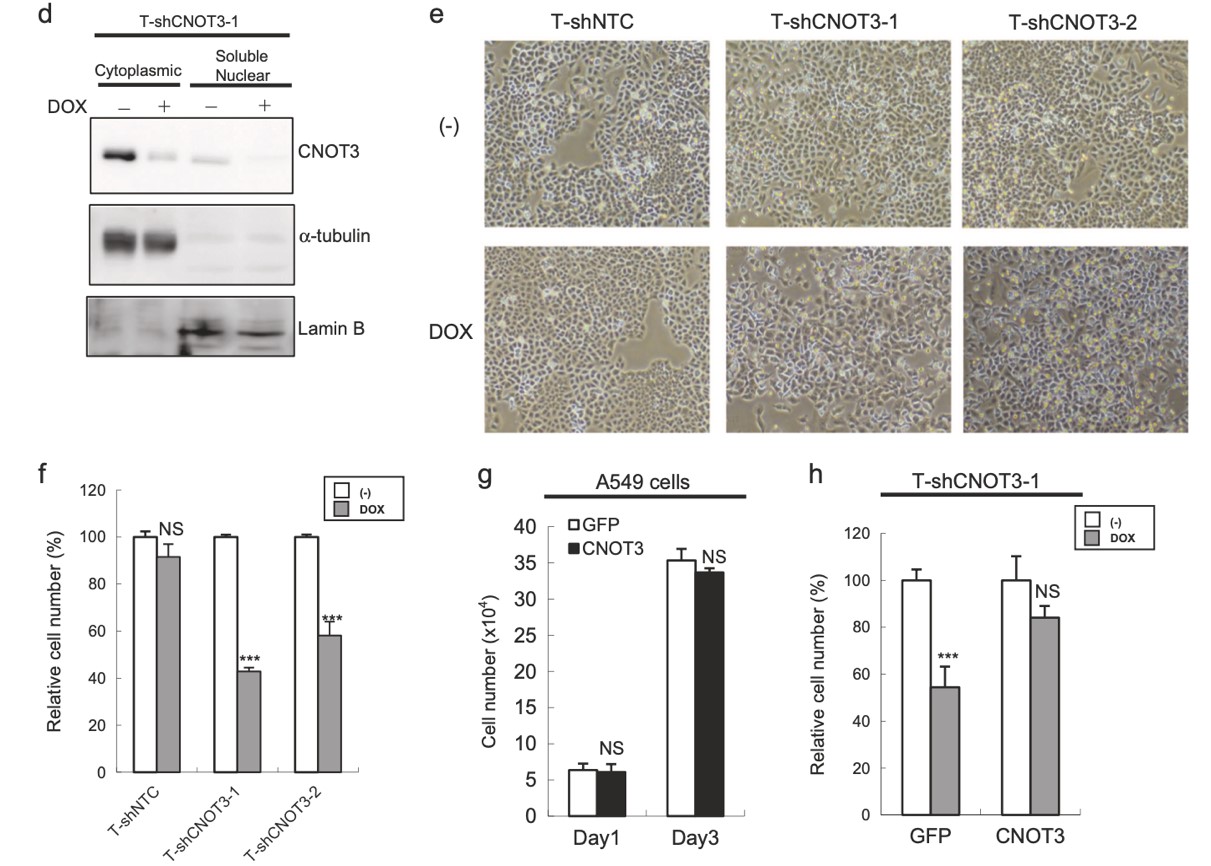
Figure 2. Depletion of CNOT3 attenuates the proliferation of human NSCLC cells. a. Subcellular fractionation of each A549 stable with or without 3 days of DOX treatment was performed to express shCNOT3. Soluble nuclear extracts and cytoplasmic extracts were subjected to immunoblotting. b. Representative pictures of A549-T-ShCNOT3-1 cells with or without 4 days of DOX treatment. c-e. Cell proliferation assay for each A549 stable. For (c) and (e), each A549 stable was treated with or without 4 days of DOX treatment.
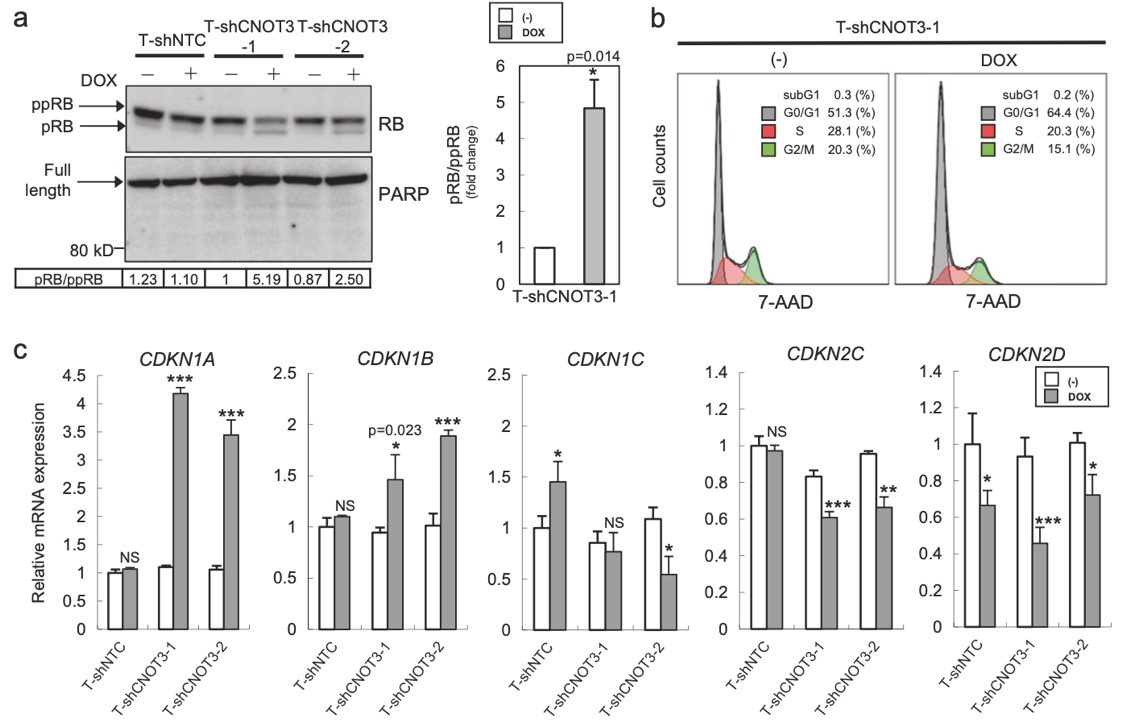
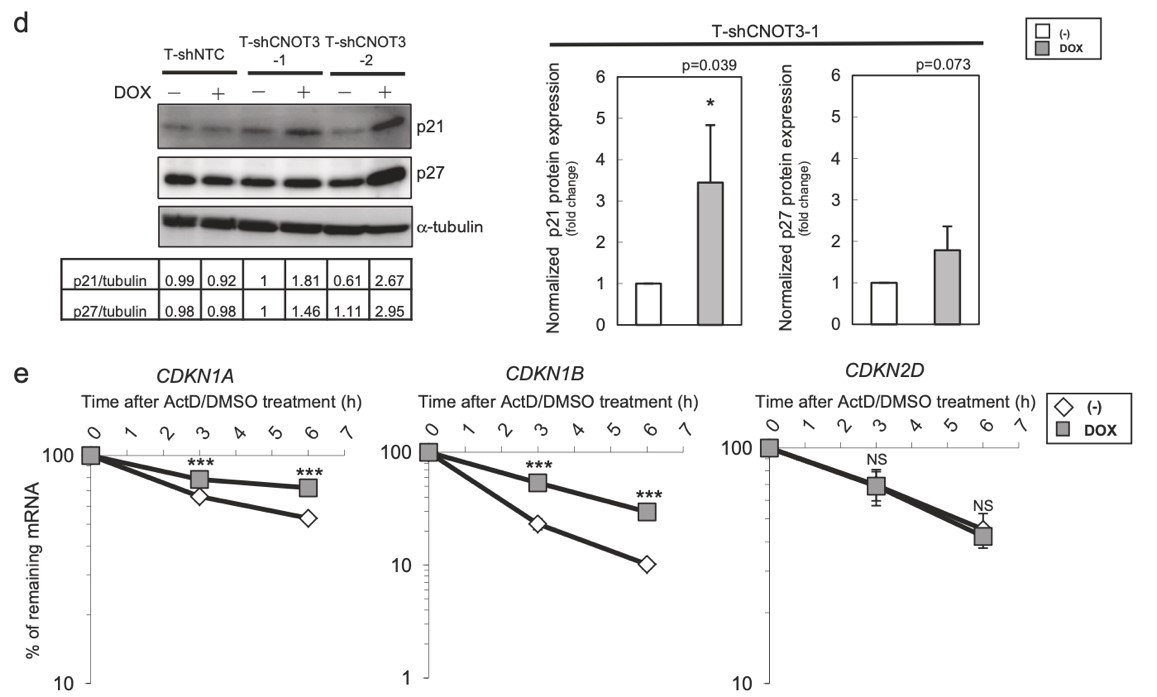
Figure 3. Depletion of CNOT3 induces the expression of p21 in human NSCLC cells. a, d Cell lysate of each A549 stable with or without 2 or 3 days of DOX treatment was subjected to immunoblotting with specific antibodies. ppRB, pRB, and full-length form of PARP are shown with arrows. Quantification of pRB/ppRB, p21 or p27/α-tubulin or β-actin was performed using Image J. The values were normalized to that of T-shCNOT3-1 without DOX. Average of the values obtained from four (p21 and p27) or three (pRB) independent sets of samples are shown. b Cell cycle analysis for A549-T-shCNOT3-1 cells with or without 3 days of DOX treatment. c qRT-PCR for CDKN1A, CDKN1B, CDKN1C, CDKN2C, and CDKN2D using the cDNA from the A549-T-shCNOT3-1 or -2 cells treated with or without DOX. e A549-T-shCNOT3-1 cells with or without 3 days of DOX treatment were treated with DMSO or ActD for 3 or 6 h. qRT-PCR for CDKN1A, CDKN1B, and CDKN2D using the cDNA from the cells with indicated time of treatment.
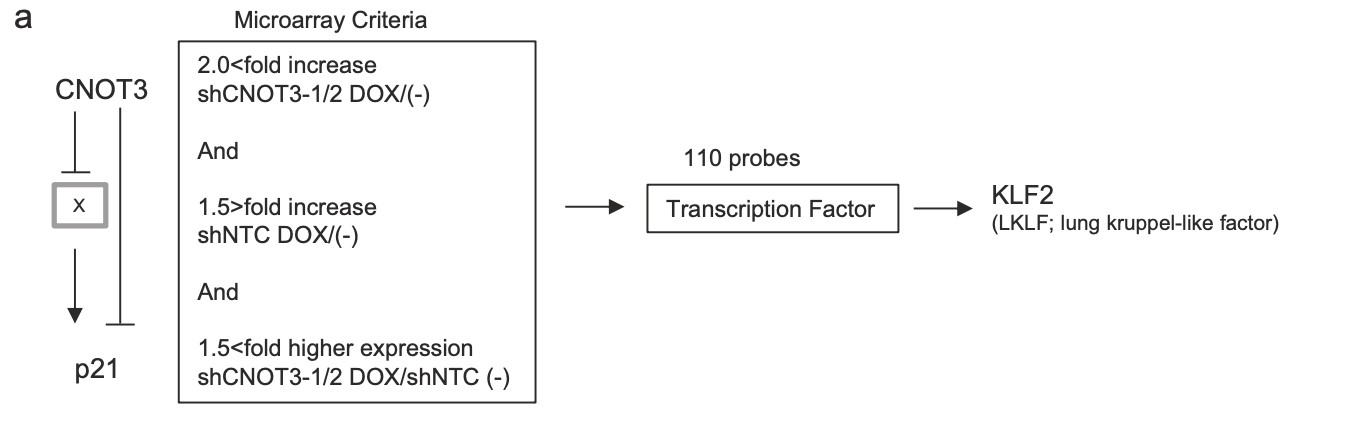
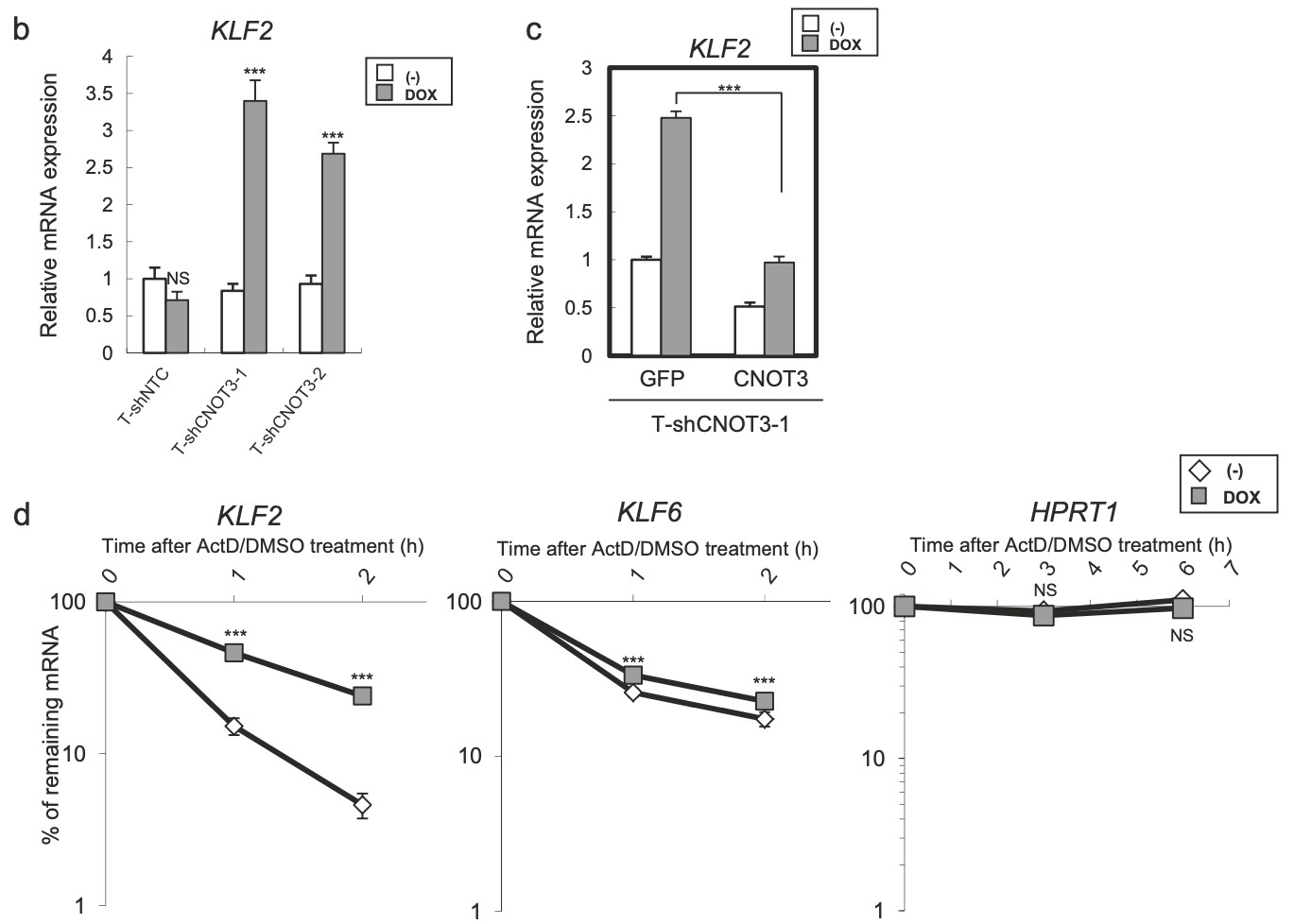
Figure 4. Identification of KLF2 transcription factor as a target of CNOT3. a Schematic model of regulation of p21 by CNOT3. We hypothesized that p21 expression is suppressed by a Factor X which is a target of CNOT3, in addition with direct mRNA decay by CNOT3. We narrowed down the candidate X to up-regulated 110 probes with microarray based on the criteria as follows: (1) More than 2-fold increase by both shCNOT3-1 and -2 induction. (2) Less than 1.5-fold increase by shNTC induction (to exclude non-specific increase). (3) More than 1.5-fold higher expression both in A549-T-shCNOT3-1 and -2 induced cells than A549-T-shNTC cells without DOX (to confirm the higher expression against A549-T-shNTC stable). We identified KLF2 as a Factor X. b qRT-PCR for KLF2 using the cDNA form the same samples with Fig. 1a. c qRT-PCR for KLF2 using the cDNA form A549-T-shCNOT3-1 cells expressing GFP or CNOT3 with or without 3 days of DOX treatment. d A549-T-shCNOT3-1 cells with or without 3 days of DOX treatment were treated with DMSO or ActD for 1 or 2 h. qRT-PCR for KLF2 and KLF6 using the cDNA from the cells with indicated time of treatment is shown. qRT-PCR for HPRT1 using the cDNA from the A549-T-shCNOT3-1 cells.
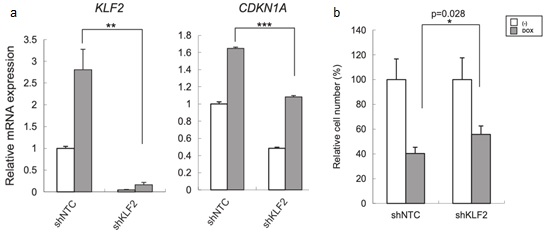
Figure 5. The growth inhibition by CNOT3 depletion is mediated by the upregulation of KLF2. a qRT-PCR for KLF2 and CDKN1A using the cDNA form A549-T-shCNOT3-1 cells expressing shNTC or shKLF2 with or without 3 days of DOX treatment. b Cell proliferation assay for each A549 cells with or without 5 days of DOX treatment. The cell number of each A549 stable without DOX was standardized to 100%.
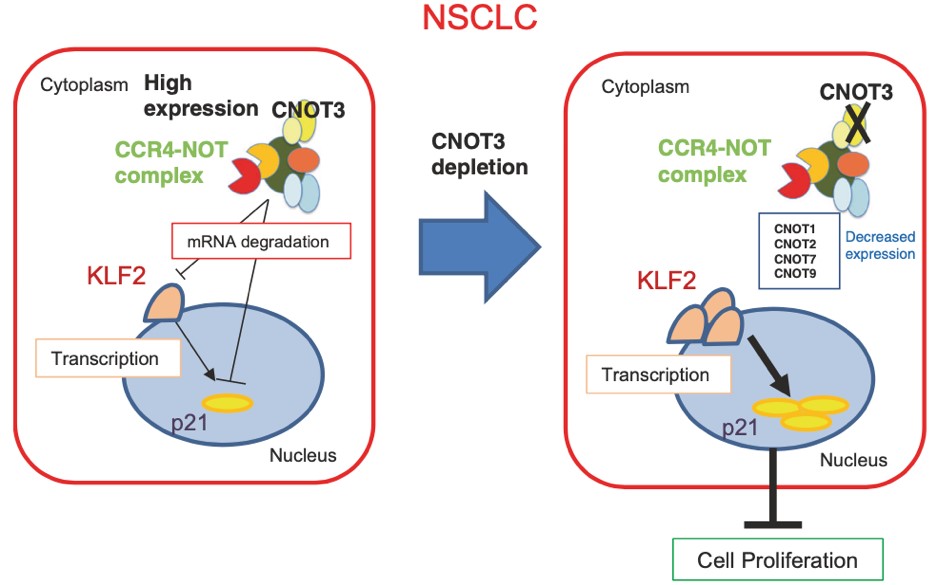
Figure 6. Schematic representation of the role of CNOT3 in NSCLC cells. CNOT3, one key subunit of the CCR4-NOT complex, is highly expressed in NSCLC cells and is required for the proper expression of some other subunits including CNOT1, CNOT2, CNOT7, and CNOT9. CNOT3 regulates the expression of p21 through mRNA degradation. CNOT3 also specifically degrades the mRNA of KLF2, which regulates the expression of p21 through transcription. When CNOT3 is depleted in NSCLC cells, the expression levels of KLF2 and p21 are elevated, resulting in the impairment of cell proliferation.
3.3 Essential functions of the CNOT7/8 catalytic subunits of the CCR4-NOT complex in mRNA regulation and cell viability.
Shortening of mRNA poly(A) tails (deadenylation) to trigger their decay is mediated mainly by the CCR4-NOT deadenylase complex. While four catalytic subunits (CNOT6, 6L 7, and 8) have been identified in the mammalian CCR4-NOT complex, their individual biological roles are not fully understood. In this study, we addressed the contribution of CNOT7/8 to viability of primary mouse embryonic fibroblasts (MEFs). We found that MEFs lacking CNOT7/8 expression [Cnot7/8-double knockout (dKO) MEFs] undergo cell death, whereas MEFs lacking CNOT6/6L expression (Cnot6/6l-dKO MEFs) remain viable. Co-immunoprecipitation analyses showed that CNOT6/6L are also absent from the CCR4-NOT complex in Cnot7/8-dKO MEFs. In contrast, either CNOT7 or CNOT8 still interacts with other subunits in the CCR4-NOT complex in Cnot6/6l-dKO MEFs. Exogenous expression of a CNOT7 mutant lacking catalytic activity in Cnot7/8-dKO MEFs cannot recover cell viability, even though CNOT6/6L exists to some extent in the CCR4-NOT complex, confirming that CNOT7/8 is essential for viability. Bulk poly(A) tail analysis revealed that mRNAs with longer poly(A) tails are more numerous in Cnot7/8-dKO MEFs than in Cnot6/6l-dKO MEFs. Consistent with elongated poly(A) tails, more mRNAs are upregulated and stabilized in Cnot7/8-dKO MEFs than in Cnot6/6l-dKO MEFs. Importantly, Cnot6/6l-dKO mice are viable and grow normally to adulthood. Taken together, the CNOT7/8 catalytic subunits are essential for deadenylation, which is necessary to maintain cell viability, whereas CNOT6/6L are not.
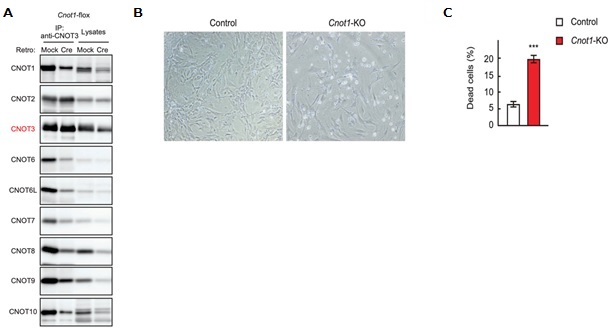
Figure 1. Cnot1-KO MEFs undergo cell death. (A) Lysates were prepared from Cnot1-flox MEFs that were infected with mock or Cre-expressing retrovirus and subjected to immunoprecipitation with anti-CNOT3 antibody. CNOT3 are shown in red to indicate a precipitated molecule. Lysates and immunoprecipitates (IP) were analysed by immunoblot with the indicated antibodies. (B) Morphology of Cnot1-flox MEFs infected with mock (Control) or Cre-expressing retrovirus (Cnot1-KO). Photographs are at the same magnification and represent one of the three independent experiments. Dead cells that were about to lose adhesion were observed in Cnot1-KO MEFs. (C) Cell death was assessed by propidium iodide uptake using flow cytometry (n = 3).
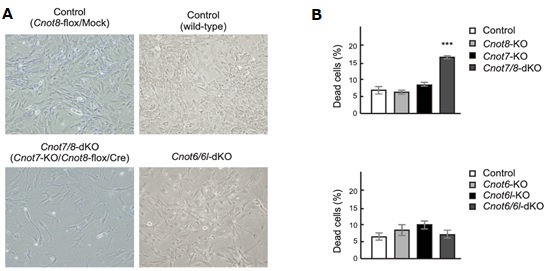
Figure 2. Suppression of CNOT7/8, but not CNOT6/6L, affects viability of MEFs. (A) Morphology of MEFs with the indicated genotypes. Mock and Cre represent cells infected with retrovirus. (B) Cell death was assessed as in Fig. 1C (n = 3).
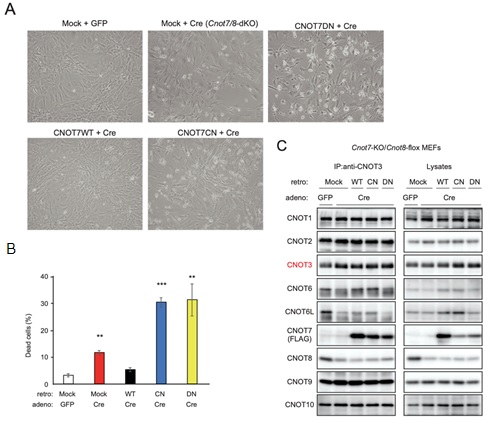
Figure 3. CNOT7 catalytic activity is sufficient to maintain MEF viability. (A) Morphology of Cnot7-KO/Cnot8-flox MEFs infected with the indicated (retro + adeno) viruses. (B) Cell death was assessed as in Fig. 1D (n = 3). (C) Lysates were prepared from Cnot7-KO/Cnot8-flox MEFs infected with the indicated viruses and subjected to immunoprecipitation with anti-CNOT3 antibody. CNOT3 is shown in red to indicate a precipitated molecule. Lysates and IP were analysed by immunoblot. WT: CNOT7 wild-type, CN: CNOT7 lacking catalytic activity, DN: CNOT7 dominant negative mutant which lacks catalytic activity and an ability to bind to CNOT6/6L.
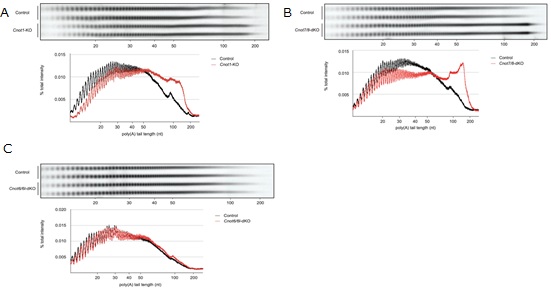
Figure 4. RNAs in Cnot1-KO and Cnot7/8-dKO MEFs have longer polyA tails compared to those in Cnot6/6l-dKO MEFs. (A-C) Poly(A) tail lengths of bulk RNA in Cnot1- KO (A), Cnot7/8-dKO (B) and Cnot6/6l-dKO MEFs (C) (n = 2 for each genotype). Cnot1-flox MEFs infected with mock retrovirus, Cnot7+/+; Cnot8-flox MEFs infected with mock retrovirus, and WT MEFs were used as controls, respectively. Densitograms of poly(A) tail lengths are shown below each image. The signal intensity normalized to total intensity (%) was calculated. The mean of two independent experiments was used.
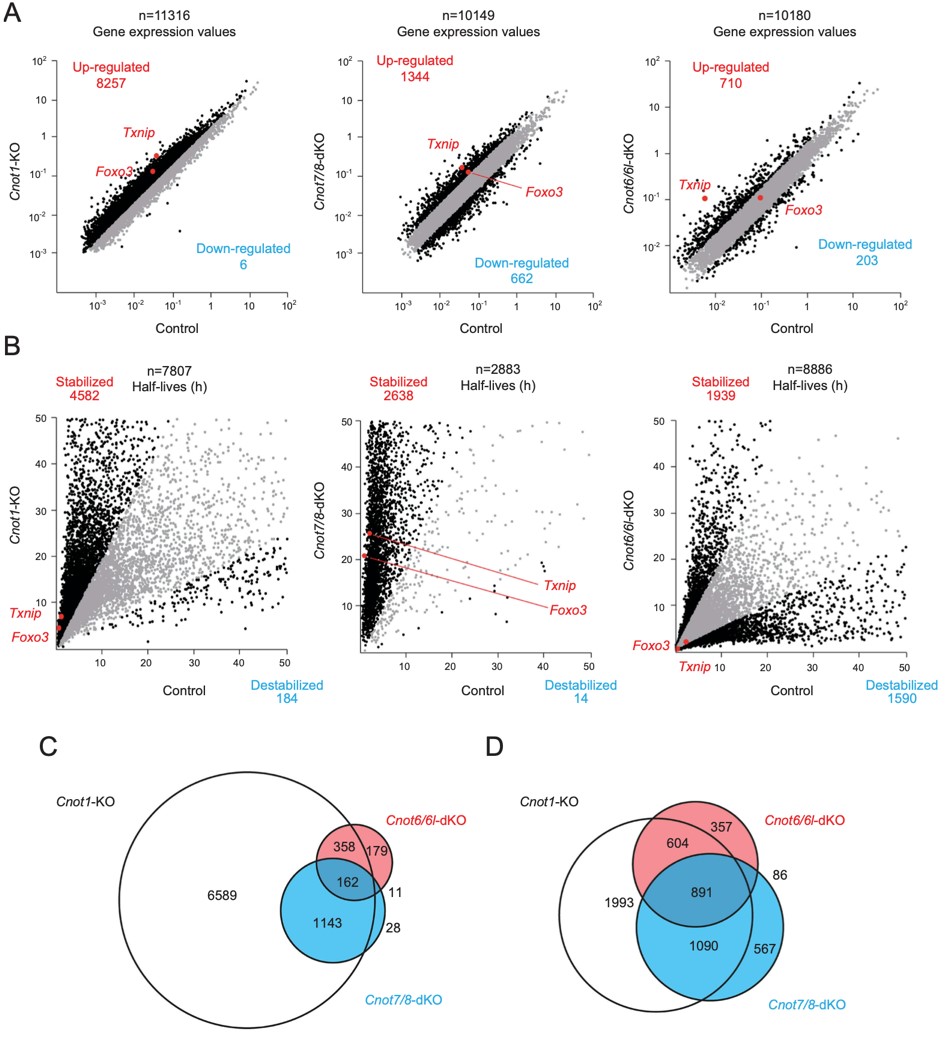
Figure 5. Gene expression and mRNA stability differ among Cnot1-KO, Cnot7/8-dKO and Cnot6/6l-dKO MEFs. RNA-seq analysis of Cnot1-KO, Cnot7/8-dKO and Cnot6/ 6l-dKO MEFs compared to controls: Cnot1-flox MEFs infected with mock retrovirus, Cnot8-flox MEFs infected with mock retrovirus and WT MEFs, respectively (n = 2). (A, B) Scatter plots comparing gene expression values (A) or mRNA half-lives (B). mRNAs showing expression (A) or half-lives (B) that differed more than twofold are displayed in black. All values represent the means of two independent experiments. (C, D) Venn diagrams showing overlap of upregulated (C) or stabilized genes (D) (more than twofold) in Cnot1-KO, Cnot7/8-dKO and Cnot6/6l-dKO MEFs compared to controls.
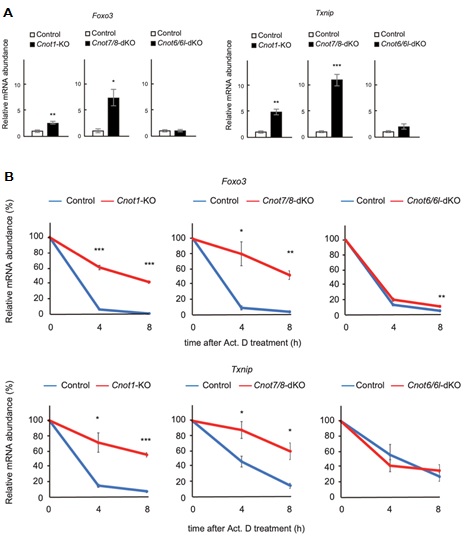
Figure 6. Foxo3 and Txnip mRNAs are upregulated and stabilized in Cnot1-KO and Cnot7/8-dKO MEFs. (A) qPCR analysis of the indicated mRNAs in Cnot1-KO, Cnot7/ 8-dKO and Cnot6/6l-dKO MEFs together with their controls, as in Fig. 5. Relative mRNA levels were determined by qPCR and normalized to the Gapdh mRNA level (n = 3). (B) Decay curves of mRNAs. Total RNAs were prepared from the indicated MEFs treated with Act. D (0, 4 or 8 h). Relative mRNA levels were determined as in (A). mRNA levels without Act. D treatment (0 h) were set to 100% (n = 3).
4. Publications
4.1 Journals
- Mostafa D, Takahashi A, Yanagiya A, Yamaguchi T, Abe T, Kureha T, Kuba K, Kanegae Y, Furuta Y, Yamamoto T, Suzuki T. Essential functions of the CNOT7/8 catalytic subunits of the CCR4-NOT complex in mRNA regulation and cell viability. RNA Biology, 17:403-416, 2020
- Nerome K, Ito-Kureha T, Paganini T, Fukuda T, Igarashi Y, Ashitomi H, Ikematsu S, Yamamoto T. Potent and broad anticancer activities of leaf extracts from Melia azedarach L. of the subtropical Okinawa islands. Am J Cancer Res, 10(2):581-594,2020
- Suzuki T, Kikuguchi C, Nishijima S, Yamamoto T. Insufficient liver maturation affects murine early postnatal hair cycle. Biochemical and Biophysical Research Communications 521:172-177, 2020
- Baba M, Yokoyama K, Seiriki K, Naka Y, Matsumura K, Kondo M, Yamamoto K, Hayashida M, Kasai A, Ago Y, Nagayasu K, Hayata-Tnaka A, Takahashi A, Yamaguchi S, Mori D, Ozaki N, Yamamoto T, Takuma K, Hashimoto R, Hashimoto H, Nakazawa T. Psychiatric-disorder-related behavioral phenotypes and cortical hyperactivity in a mouse model of 3q29 deletion syndrome. Neuropsycopharacology 44:2125-2135, 2019
- Takahashi A, Takaoka S, Kobori S, Yamaguchi T, Ferwati S, Kuba K, Yamamoto T, Suzuki T. The CCR4-NOT deanylase complex maintains adipocyte identity. Int. J. Mol. Sci. 20: 5274, 2019.
- Montrose K, Kobayashi S, Manabe T, Yamamoto T. Lmtk3-KO mice display a range of behavioural abnormalities and have an impairment in GluA1 trafficking. Neuroscience 414:154-167, 2019
- Ashworth W, Stoney P, Yamamoto T. States of Decay: The Systems Biology of mRNA Stability. Current Opinion in Systems Biology 15:48-57, 2019.
- Morita M, Siddiqui N, Katsumura S, Rouya C, Larsson O, Nakashima T, Hekmatnejad B, Takahashi A, Kiyonari H, Zang M, St-Arnaud R, Oike Y, Giguère V, Topisirovic I, Okada-Hatakeyama M, Yamamoto T, Sonenberg N. Hepatic post-transcriptional network comprising of CCR4-NOT deadenylase and FGF21 maintains systemic metabolic homeostasis. Proc Nat Acad Sci USA 116:7973-7981, 2019
- Shirai Y, Mizutani A, Nishijima S, Horie M, Kikuguchi C, Elisseeva O, Yamamoto T. CNOT3 targets negative cell cycle regulators in non-small cell lung cancer development. Oncogene 38:2580-2594, 2019
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- P. Stoney, A. Yanagiya, T. Yamamoto. Upregulation of Cnot8 in Cnot7 KO cells to maintain poly(A) homeostasis. The 42nd Annual Meeting of the Molecular Biology Society of Japan, Fukuoka, Japan, 2019.12.06 (2019)
- S. Nishijima, T. Suzuki, T. Yamamoto. Suppression of CNOT11, a subunit of CCR4-NOT complex, induces cell cycle arrest and autophagy. The 42nd Annual Meeting of the Molecular Biology Society of Japan, Fukuoka, Japan, 2019.12.06 (2019)
- W. Ashworth, Y. Yamamoto. Multi-omics analysis of mammalian CNOT4. The 42nd Annual Meeting of the Molecular Biology Society of Japan, Fukuoka, Japan, 2019.12.06 (2019)
- A. Yanagiya, T. Yamamoto. Post-transcriptional regulation in insulin biosynthesis by the Ccr4-Not deadenylase complex in mouse pancreatic β cells. The 42nd Annual Meeting of the Molecular Biology Society of Japan, Fukuoka, Japan, 2019.12.05 (2019)
- T. Yamamoto. Introduction to IMS and to my Research. Riken-KU Leuven Joint Symposium, KU-Leuven, Brussels, 2019.12.04 (2019)
- S. Soeda, T. Yamamoto. The role of maternal mRNA regulation by CCR4-NOT complex during mouse early development. The 42nd Annual Meeting of the Molecular Biology Society of Japan, Fukuoka, Japan, 2019.12.03 (2019)
- S. Takaoka, A. Takahashi, H. MA Mohamed, P. Stoney, T. Yamamoto. Brain-specific knock-out of 5’ - 3’ exoribonuclease Xrn1 causes hyperphagia and obesity. The 26th East Asia Joint Symposium on Biomedical Research, Seoul, South Korea, 2019.10.24 (2019)
- S. Soeda, T. Yamamoto. The role of gene expression regulation by CCR4-NOT mRNA deadenylation complex during mouse early development. RNA Frontier Meeting 2019, Shizuoka, Japan, 2019.09.24 (2019)
- M. Youssef, Y. Kiyama, H. Hamada, T. Suzuki, T. Manabe, T. Yamamoto. Transducer of ErbB2 (Tob) regulates stress in the brain. IBRO 2020, Daegu, South Korea, 2019.09.23 (2019)
- T. Yamamoto. Studies on breast cancer genes: history and latest trend. Okinawa Breast Cancer Research Association, Okinawa, Japan, 2019.08.24 (2019)
- K. Matsuura, T. Yamamoto. Investigation of the role of CNOT9 in mature brain. The 7th CCR4-NOT Meeting, Miyagi, Japan, 2019.07.27 (2019)
- P. Stoney, A. Yanagiya, T. Yamamoto. Upregulation of Cnot8 in Cnot7 KO cells to maintain poly(A) homeostasis. The 7th CCR4-NOT Meeting, Miyagi, Japan, 2019.07.27 (2019)
- H. Sarmah, T. Yamamoto. Role of mammalian CNOT9 in early embryonic development and differentiation. The 7th CCR4-NOT Meeting, Miyagi, Japan, 2019.07.27 (2019)
- S. Takaoka, A. Takahashi, H. MA Mohamed, P. Stoney, T. Yamamoto. Brain-specific knock-out of 5’ - 3’ exoribonuclease Xrn1 causes hyperphagia and obesity. The 7th CCR4-NOT Meeting, Miyagi, Japan, 2019.07.27 (2019)
- A. Yanagiya, T. Yamamoto. Post-transcriptional regulation maintains redox homeostasis in insulin biosynthesis by the Ccr4-Not deadenylase complex. The 7th CCR4-NOT Meeting, Miyagi, Japan, 2019.07.26 (2019)
- M. Youssef, Y. Kiyama, H. Hamada, K. Montrose, P. Stoney, T. Suzuki, T. Manabe, T. Role of Tob in the brain: An insight into stress-responsiveness function. Japanese Neuroscience society (Neuro2019), Niigata, Japan, 2019.07.25 (2019)
- A. Yanagiya, T. Yamamoto. Post-transcriptional regulation maintains redox homeostasis in insulin biosynthesis by the Ccr4-Not deadenylase complex. The 21st annual meeting of the RNA society of Japan, Tokyo, Japan, 2019.07.18 (2019)
- A. Yanagiya, T. Yamamoto. Post-transcriptional regulation to maintain redox homeostasis in insulin biosynthesis by the Ccr4-Not deadenylase complex in mouse pancreatic β-cells. mRNA Turnover Mechanisms, Regulation and their Implication in Infectious and Age-Related Diseases, Montreal, Canada, 2019.06.27 (2019)
- S. Soeda, K. Yamada-Nomoto, T. Michiue, M. Ohsugi. RSK-MASTL pathway delays meiotic exit in mouse zygotes to ensure paternal chromosome stability. The Annual Meeting of Japanese Society of Cell Science, Kobe, Japan, 2019.06.24 (2019)
- T. Yamamoto. Disease systems biology research in IMS and my cell signal study. RIKEN Center for Advanced Photonics, Riken, Wako, Japan, 2019.05.17 (2019)
- G. Vares, Y. Saintigny, F. Chevalier, C. Lepleux, M. Temelie, H. Hoh, V. Jallet, S. Sai, H. Sugawara, T. Nakajima. Targeting cancer stem cells with miRNA-based strategies and particle radiation therapy. HIMAC Annual Meeting, Chiba, Japan, 2019.04.22 (2019)
- G. Vares, Y. Saintigny, S. Sai, T. Nakajima. Characterizing and targeting cancer stem cells in challenging cancers. International Congress of Radiation Research (ICRR), Manchester, UK, 2019.08.26 (2019)
5. Intellectual Property Rights and Other Specific Achievements
5.1 Intellectual Property Rights
Title: Anti-tumor agent
Patent Number: JP6443872B2
Inventors: Kuniaki Nerome, Tadashi Yamamoto, Taku Kureha, Reiko Nerome
Owner: The Institute of Biological Resources
Priority Date: Nov. 10, 2016
6. Meetings and Events
6.1 Seminar:
6.1.1 Heteromeric interference, a novel pathogenesis for human immunodeficiency
- Date: April 18, 2019
- Venue: OIST Campus Lab1
- Speaker: Dr. Ichiro Taniuchi (RIKEN, Yokohama)
6.1.2 Mechanisms determine gene silencing efficacies by miRNAs/siRNAs: thermodynamic properties and secondary structures.
- Date: May 8, 2019
- Venue: OIST Campus Lab1
- Speaker: Dr. Kumiko Ui-Tei (The University of Tokyo)
6.1.3 Decoding environmentally driven gene regulatory networks in hepatic macrophages
- Date: December 10, 2019
- Venue: OIST Campus Lab1
- Speaker: Dr. Mashito Sakai (The University of California, San Diego)
6.1.4 A hepatic post-transcriptional control of whole body metabolic homeostasis through FGF21 regulation by CCR4-NOT deadenylase
- Date: December 11, 2019
- Venue: OIST Campus Lab1
- Speaker: Dr. Sakie Katsumura (The University of Texas)
6.1.5 The reason why miRNAs bind to CDS
- Date: December 13, 2019
- Venue: OIST Campus Lab1
- Speaker: Dr. Hiroaki Sako (The University of Tokyo)
6.1.6 Portrait of 40-year-old p53
- Date: January 22, 2020
- Venue: OIST Campus Lab1
- Speaker: Dr. Rieko Ohki (National Cancer Center Research Institute)
6.1.7 Starting from the ends: from eukaryotic mRNA decay mechanisms to diseases
- Date: January 22, 2020
- Venue: OIST Campus Lab1
- Speaker: Dr. Bertrand Séraphin (University of Strasbourg)
6.1.8 The Not proteins regulate translation elongation dynamics
- Date: January 23, 2020
- Venue: OIST Campus Lab1
- Speaker: Dr. Martine Collart (The University of Geneva)
6.1.9 High throughput screening systems for inhibitors of phosphorylation dependent signaling molecules using fluorescent proteins
- Date: January 29, 2020
- Venue: OIST Campus Lab1
- Speaker: Dr. Nobumoto Watanabe (RIKEN, Wako)
6.1.10 How Aurora B activity is controlled at centromeres in mitosis
- Date: February 5, 2020
- Venue: OIST Campus Lab1
- Speaker: Dr. Toru Hirota (Cancer Institute of the Japanese Foundation for Cancer Research (JFCR))
6.1.11 Long non-coding RNAs required for MYC-driven cell proliferation
- Date: February 12, 2020
- Venue: OIST Campus Lab1
- Speaker: Dr. Peter K. Vogt (Scripps Research Institute)
6.1.12 Colon cancer metastasis: from mouse models to clinical applications
- Date: February 19, 2019
- Venue: OIST Campus Lab1
- Speaker: Dr. Makoto Taketo (Graduate School of Medicine, Kyoto University)
6.1.13 Transcription/export complex (TREX), elongation rate and mRNA 3´end processing
- Date: February 27, 2020
- Venue: OIST Campus Lab1
- Speaker: Dr. Teruko Tamura-Neimann (Hannover Medical School)
6.1.14 Cancer stem cells: targets for cancer eradication
- Date: Mar 18, 2020
- Venue: OIST Campus Lab1
- Speaker: Dr. Hideyuki Saya (Keio University School of Medicine)
6.1.15 Regulation of T cell activation and function by Innate signaling
- Date: Mar 25, 2020
- Venue: OIST Campus Lab1
- Speaker: Dr. Takashi Saito (RIKEN, Yokohama)
6.2 Meeting
6.2.1 ICSB 2019 (in collaboration with DoR)
- Date: November 1-5, 2019
- Venue: OIST Auditorium and Meeting Rooms
- Organizer (Core members): Shinya Kuroda (University of Tokyo), Miki Ebisuya (EMBL), Mariko Okada (Osaka University), Atsushi Mochizuki (Kyoto University), Katsuyuki Yugi (RIKEN), Kenichi Hironaka (University of Tokyo), Yohei Morita (OIST), Tomomi Nishi (OIST), Mayumi Kudo (JST), Haruka Ikuta (JST), Tadashi Yamamoto (OIST)
- The number of Participants: 442
6.2.2 The State-of-the-Art 3D Tissue Culture & Organoids Symposium (in collaboration with DoR)
- Date: April 18-20, 2019
- Venue: OIST Seaside House.
- Organizer: Keiko Kono (OIST), Mitsuru Morimoto (RIKEN), Toshiro Sato (Keio University), Minoru Takasato (RIKEN), Tadashi Yamamoto (OIST)
- The number of Participants: 30
7. Other
Nothing to report.



