FY2016 Annual Report
Cell Signal Unit
Professor Tadashi Yamamoto
Abstract
The Cell Signal Unit studies various molecular and cellular events that are important for maintaining healthy life in response to environments. The Unit explores the cause of various diseases that include cancer, neuronal disorder, immunological diseases, and diabetes/obesity at the molecular level. To approach these issues, the Unit characterizes regulation of gene expression at the level of RNA that includes micro RNA, mRNA and long non-coding RNA. The Unit also characterizes cell signaling with special interest on control of the brain function such as emotions, learning and memory.
1. Staff
- Dr. Toru Suzuki, Group Leader
- Dr. Olga Elisseeva, Group Leader
- Dr. Patrick Stoney, Staff Scientist
- Dr. Ken Matsuura, Staff Scientist
- Dr. Yo-taro Shirai, Postdoctoral Scholar
- Dr. Taku Kureha, Postdoctoral Scholar
- Dr. Akinori Takahashi, Postdoctoral Scholar
- Dr. Kristopher Paraiso Montrose, Postdoctoral Scholar
- Ms. Chisato Kikuguchi, Technical Staff
- Ms. Miho Tokumasu, Technical Staff
- Ms. Tiziana Paganini, Technical Staff
- Ms. Saori Nishijima, Technical Staff
- Ms. Risa Ishida, Technical Staff
- Ms. Sandrine Anne Laure Burriel, Graduate Student
- Mr. Haytham Mohamed Aly Mohamed, Graduate Student
- Mr. Shohei Takaoka, Graduate Student
- Mr. Hemanta Sarmah, Graduate Student
- Ms. Dina Mostafa, Graduate Student
- Mr. Mohieldin Magdy Mahmoud Youssef, Graduate Student
- Mr. Hong Huat Hoh, Graduate Student
- Ms. Kaori Yamashiro, Research Unit Administrator
2. Collaborations
2.1 Physiological studies of the CCR4-NOT complex
• Description: Analyze the physiological role of each component of the CCR4-NOT complex using gene-modified mice
• Type of collaboration: Joint research
• Researchers: Kuba K. Department of Physiology, Graduate School of Medicine, Akita University
2.2 Basic cancer research for prevention and treatment
• Description: Search for substances that contribute to the prevention and treatment of cancer from biological resources, and elucidate its mechanism of action
• Type of collaboration: Joint researches I and II
• Researchers:
o For I: Representative director Kuniaki Nerome, Bioresource Laboratory LLC.
o For II: Professor Shinya Ikematsu, National Institute of Technology, Okinawa College
2.3 In vivo histology and behavioral analysis of psychiatric model mice
• Description: Analysis of brain structure of 3q29 CNV mice with the state-of-the-art 11.7-Tesla MRI as well as investigating behavior of the mice
• Type of collaboration: Joint research
• Researchers: Dr. Kazumasa Yokoyama, Takeda Pharmaceutical Company Ltd.
Dr. Takanobu Nakazawa, School of Pharmaceutical Science, Osaka University
2.4 Electrophysiology
• Description: Electrophysiological studies of gene modified mice with abnormal social behavior
• Type of collaboration: Joint research
• Researchers: Professor Toshiya Manabe, Institute of Medical Science, University of Tokyo
3. Activities and Findings
3.1 The CCR4-NOT deadenylase activity contributes to generation of induced pluripotent stem cells.
Somatic cells can be reprogrammed as induced pluripotent stem cells (iPSCs) by introduction of the transcription factors, OCT3/4, KLF4, SOX2, and c-MYC. The CCR4-NOT complex is the major deadenylase in eukaryotes. Its subunits Cnot1, Cnot2, and Cnot3 maintain pluripotency and self-renewal of mouse and human embryonic stem (ES) cells and contribute to the transition from partial to full iPSCs. However, little is known about how the CCR4-NOT complex post-transcriptionally regulates the reprogramming process. Here, we show that the CCR4-NOT deadenylase subunits Cnot6, Cnot6l, Cnot7, and Cnot8, participate in regulating iPSC generation. Cnot1 knockdown suppresses expression levels of Cnot6, Cnot6l, Cnot7, and Cnot8 in mouse embryonic fibroblasts (MEFs) and decreases the number of alkaline phosphatase (ALP)-positive colonies after iPSC induction. Intriguingly, Cnot1 depletion allows Eomes and p21 mRNAs to persist, increasing their expression levels. Both mRNAs have longer poly(A) tails in Cnot1-depleted cells. Conversely, forced expression of a combination of Cnot6, Cnot6l, Cnot7, and Cnot8 increases the number of ALP-positive colonies after iPSC induction and decreases expression levels of Eomes and p21 mRNAs. Based on these observations, we propose that the CCR4-NOT deadenylase activity contributes to iPSC induction.
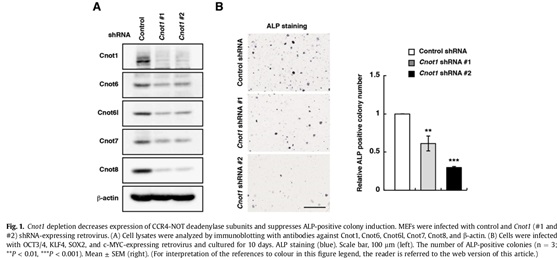
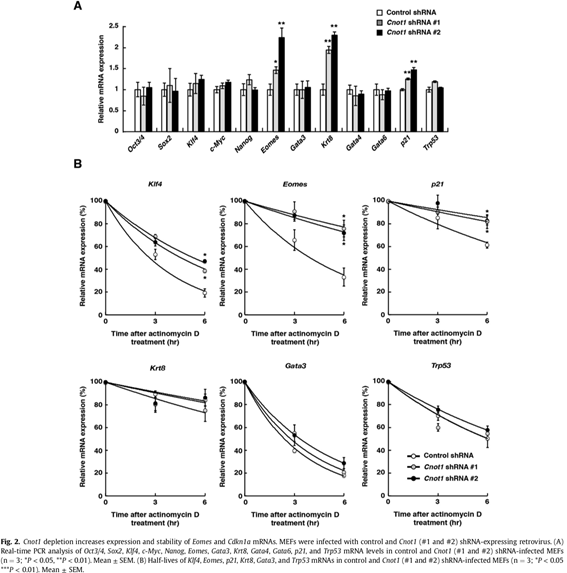
3.2 Structural basis for inhibition of the deadenylase activity of human CNOT6L.
Human CNOT6L/CCR4, a member of the endonuclease-exonuclease-phosphatase (EEP) family enzymes, is one of the two deadenylase enzymes in the conserved CCR4-NOT complex. Here, we report inhibitor-bound crystal structures of the human CNOT6L nuclease domain in complex with the nucleotide CMP and the aminoglycoside neomycin. Deadenylase activity assays show that nucleotides are effective inhibitors of both CNOT6L and CNOT7, with AMP more effective than other nucleotides, and that neomycin is a weak deadenylase inhibitor. Structural analysis shows that all inhibitors occupy the substrate and magnesium-binding sites of CNOT6L, suggesting that inhibitors compete with both substrate and divalent magnesium ions for overlapping binding sites.
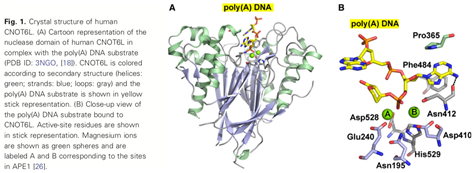

3.3 Emerging roles of ARHGAP33 in intracellular trafficking of TrkB and pathophysiology of neuropsychiatric disorders
Intracellular trafficking of receptor proteins is essential for neurons to detect various extracellular factors during the formation and refinement of neural circuits. However, the precise mechanisms underlying the trafficking of neurotrophin receptors to synapses remain elusive. Here, we demonstrate that a brain-enriched sorting nexin, ARHGAP33, is a new type of regulator for the intracellular trafficking of TrkB, a high-affinity receptor for brain-derived neurotrophic factor. ARHGAP33 knockout (KO) mice exhibit reduced expression of synaptic TrkB, impaired spine development and neuropsychiatric disorder-related behavioural abnormalities. These deficits are rescued by specific pharmacological enhancement of TrkB signalling in ARHGAP33 KO mice. Mechanistically, ARHGAP33 interacts with SORT1 to cooperatively regulate TrkB trafficking. Human ARHGAP33 is associated with brain phenotypes and reduced SORT1 expression is found in patients with schizophrenia. We propose that ARHGAP33/SORT1-mediated TrkB trafficking is essential for synapse development and that the dysfunction of this mechanism may be a new molecular pathology of neuropsychiatric disorders.
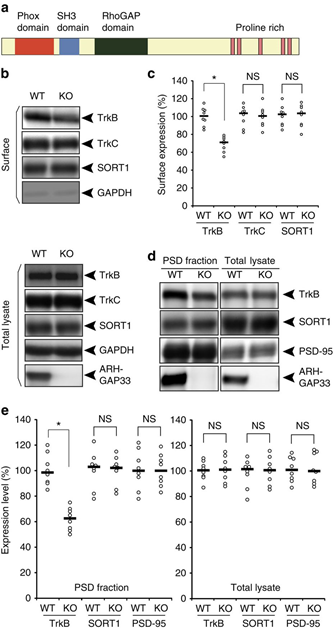
(a) Protein structure of a brain-enriched SNX protein, ARHGAP33. ARHGAP33 has an N-terminal PX domain, an SH3 domain and a RhoGAP domain. (b,c) Decreased cell-surface expression of TrkB in ARHGAP33 KO mice. Biotinylated cell-surface proteins (upper) and total lysates (lower) of WT and ARHGAP33 KO neurons (14 DIV) were immunoblotted with anti-TrkB, anti-TrkC, anti-SORT1, anti-GAPDH and anti-ARHGAP33 antibodies. (b) Representative blots. (c) Quantification of surface expression (each, n=8; TrkB, P=7.8 × 10−4; TrkC and SORT1, P>0.05; Mann–Whitney U-test). The expression levels of TrkB, TrkC and SORT1 in ARHGAP33 KO neurons were normalized to those in WT neurons (The averaged WT values were set to 100%). (d,e) Decreased TrkB in the isolated PSD fraction of ARHGAP33 KO mice. The isolated PSD fraction and total lysates of WT and ARHGAP33 KO mice were immunoblotted with anti-TrkB, anti-PSD-95, anti-SORT1, and anti-ARHGAP33 antibodies. Representative blots (d). Quantification for the isolated PSD fraction (each, n=8, TrkB, P=7.7 × 10−4; SORT1 and PSD-95, P>0.05; Mann–Whitney U-test) and for the total lysate (each, n=8, P>0.05; Mann–Whitney U-test; e) The expression levels of TrkB, SORT1 and PSD-95 in the PSD fraction and total lysate from ARHGAP33 KO mice were normalized to those from WT mice (The averaged WT values were set to 100%). Note that the amounts of PSD-95 and SORT1 in the isolated PSD fraction from ARHGAP33 KO mice were not significantly different from those in the fraction from WT mice. *P<0.05. NS, not significant. Bars show median values. All western blots show representative results from eight independent experiments performed using different mice.
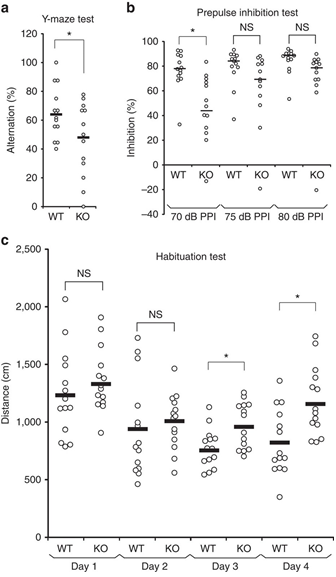
(a) Impaired spontaneous alternations of ARHGAP33 KO mice during the Y-maze test (WT, n=14, KO, n=13, F1,25=5.18, P=0.031, one-way ANOVA). *P<0.05. Bars show mean values. (b) Impaired PPI of ARHGAP33 KO mice (each n=13, P=0.046, Friedman test followed by Scheffe tests). *P<0.05. NS, not significant. Bars show median values. (c) Impaired open field habituation of ARHGAP33 KO mice during the openfield habituation test (each n=14, genotype effect, F1,26=5.08, P=0.033, two-way ANOVA with repeated measures; day 3, P=1.3 × 10−4, day 4, P=7.0 × 10−4, Tukey–Kramer post hoc tests). *P<0.05. NS, not significant. Bars show mean values.
4. Publications
4.1 Journals
- Tsai H, Cheng J, Chang H, Yamamoto T and Shen A. Uniform electric field generation in circular multi-well culture plates using polymeric inserts. Sci Rep 6:26222,2016
- Katano T, Fukuda M, Furue H, Yamazaki M, Abe M, Watanabe M, Nishida K, Yao I, Yamada A, Hata Y, Okumura N, Yamamoto T, Sakimura K, Takao T and Ito S. Involvement of brain-enriched guanylate kinase-associated protein (BEGAIN) in chronic pain after peripheral nerve injury. eNeuro 10.1523/ENEURO.0110-16,2016
- Ban T, Sato G R ,Nishiyama A, Akiyama A, Takasuna M, Umehara M, Suzuki S, Ichino M, Matsunaga S, Kimur A, Kimura Y, Yanai H, Miyashita S, Kuromitsu J, Tsukahara K,Yoshimatsu K, Endo I, Yamamoto T, Hirano H, Ryo A, Taniguchi T and Tamura T. Lyn Kinase Suppresses the Transcriptional Activity of IRF5 in the TLR-MyD88 Pathway to Restrain the Development of Autoimmunity. Immunity 45(2):319-32,2016
- Ohashi T, Yamamoto T, Yamanashi Y and Ohsugi M. Human TUBG2 gene is expressed as two splice variant mRNAs and involved in cell growth. FEBS 590(8):1053-63,2016
- Zhang Q, Yan D, Guo E, Ding B, Yang W, Liu R, Yamamoto T and Bartlam M. Structural basis for inhibition of the deadenylase activity of human CNOT6L. FEBS letters 590(8):1270-9,2016
- Zukeran A, Takahashi A, Takaoka S, Mohamed H, Suzuki T, Ikematsu S and Yamamoto T. The CCR4-NOT deadenylase activity contributes to generation of induced pluripotent stem cells. BBRC 474(2):233-9,2016
- Nakazawa T, Hashimoto R, Sakoori K, Sugaya Y, Tanimura A, Hashimotodani Y, Ohi K, Yamamori H, Yasuda Y, Umeda-Yano S, Kiyama Y, Konno K, Inoue T, Yokoyama K, Inoue T, Numata S, Ohnuma T, Iwata N, Ozaki N, Hashimoto H, Watanabe M, Manabe T, Yamamoto T, Takeda M and Kano M. Emerging Roles of ARHGAP33 in Intracellular Trafficking of TrkB and Pathophysiology of Neuropsychiatric Disorders. Nat Commun 7:10594, 2016
- Li T, Amari T, Semba K, Yamamoto T and Takeoka S. Construction and evaluation of pH-sensitive immunoliposomes for enhanced delivery of anticancer drug to ErbB2 over-expressing breast cancer cells. Nanomedicine 13(3):1219-1227,2017
- Li X, Morita M, Kikuguchi C, Takahashi A, Suzuki T and Yamamoto T. Adipocyte-specific disruption of mouse Cnot3 causes lipodystrophy. FEBS Letters 591(2):358-368,2017
- Chang H, Hoshina N, Zhang C, Ma Y, Cao H, Wang Y, Wu D, Bergen E, Landén M, Hultman C, Preisig M, Kutalik Z, Castelao E, Grigoroiu-Serbanescu M, Forstner A, Strohmaier J, Hecker J, Schulze T, Müller-Myhsok B, Reif A, Mitchell P, Martin N, Cichon S, Nöthen M, The Swedish Bipolar Study Group, MooDS Bipolar Consortium, Walter H, Erk S, Heinz A, Meyer-Lindenberg A, Tost H, Xiao X, Yamamoto T, Rietschel M and Li M. The Protocadherin 17 Gene Affects Cognition, Personality, Amygdala Structure and Function, Synapse Development and Risk of Major Mood Disorders. Mol Psychiatry 00:1-13,2017
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- Kureha, T. (2016). Deadenylase-mediated poly(A) shortening regulates the maintenance of regulatory T cells. The 2nd Meeting of Osteoimmunology. Hotel Monterey Okinawa Spa & Resort.
- Kureha, T. (2016). Deadenylase-mediated poly(A) tail shortening regulates the positive selection of thymocytes through post-transcriptional down-regulation of ASK1. 46th Annual Meeting German Society for Immunology, Hamburg, Germany.
- Kureha, T. (2016). Poly(A) shortening of ASK1 mRNA contributes to positive selection of thymocytes by impairing TCR-induced stress response. The 8th Sinal Network Conference Research Institute for Microbial Diseases, Osaka University.
- Kureha, T. (2016). Poly(A) shortening of ASK1 mRNA contributes to positive selection of thymocytes through impairment of TCR-induced stress response. International Congress of Immunology 2016. Melbourne.
- Kureha, T. (2017). Roles of the CCR4-NOT complex in T cell differentiation and cancer immune regulation. OIST, The 5th CCR4-NOT Meeting.
- Matsuura, K. (2016). SPAL1 interacts with the Neurabin family of proteins and is involved in the regulation of G Protein-Coupled Receptor signaling. Pacifico Yokohama, 39th Annual meeting of the molecular biology society of Japan.
- Mohamed, H. (2017). CNOT1 regulation of eukaryotic circadian rhythm through post-transcriptional mechanisms. OIST, The 5th CCR4-NOT Meeting.
- Mohamed, H., et al. (2016). CNOT1 regulates eukaryotic circadian molecular clock through post-transcriptional mechanisms. Pacifico Yokohama, 39th Annual meeting of the molecular biology society of Japan.
- Montrose, K. (2016). Lmtk3 knockout mice exhibit schizophrenic-like behaviour. Hawaii, USA, 4th Annual Molecular Pyschiatry Meeting.
- Montrose, K. and T. Yamamoto (2016). Lmtk3 knockout mice exhibit schizophrenic-like behaviour. Pacifico Yokohama, 39th Annual meeting of the molecular biology society of Japan.
- Sarma, H. (2017). Physiological and Molecular basis of CNOT9. OIST, The 5th CCR4-NOT Meeting.
- Sarmah, H., et al. (2016). Role of CNOT9 in early T-cell development. Pacifico Yokohama, 39th Annual meeting of the molecular biology society of Japan.
- Shirai, Y.-t., et al. (2016). Functional analysis of CNOT3 in non-small cell lung cancer. Pacifico Yokohama, The 75th Annual Meeting of the Japanese Cancer Association.
- Suzuki, T., et al. (2016). Pacifico Yokohama, 39th Annual meeting of the molecular biology society of Japan.
- Suzuki, T., et al. (2016). CNOT6 and 6L are dispensable for keeping viability of MEFs and liver function. RNA2016, ICC Kyoto (Kyoto International Conference Center).
- Takahashi, A. (2017). Deadenylation regulates mRNA stability and transcription activity. OIST, The 5th CCR4-NOT Meeting.
- Takahashi, A., et al. (2016). Obesity and mRNA decay. RNA2016, ICC Kyoto (Kyoto International Conference Center).
- Takahashi, A., et al. (2016). Poly(A) tail length-dependent regulation of mRNA decay rate and transcription activity. Pacifico Yokohama, 39th Annual meeting of the molecular biology society of Japan.
- Takaoka, S. (2017). Feedback control of transcription by mRNA decay factor. OIST, The 5th CCR4-NOT Meeting.
- Takaoka, S., et al. (2016). Mathematical modeling of deadenylation dependent mRNA decay. ICSB 2016. Barcelona, Spain.
- Takaoka, S., et al. (2016). Pacifico Yokohama, 39th Annual meeting of the molecular biology society of Japan.
- Yamamoto, T. (2016). The CCR4-NOT deadenylase: a key player of novel paradigm of gene regulation. Institute for Advanced Biosciences, Keio University.
- Yamamoto, T. (2016). mRNA biology in life. RNA Granule. National Istitute for Basic Biology.
- Yamamoto, T. (2016). mRNA Biology -The CCR4-NOT deadenylase: a key player of novel paradigm of gene regulation-. RIKEN Center for Integrative Medical Sciences.
- Yamamoto, T., et al. (2016). Pacifico Yokohama, 39th Annual meeting of the molecular biology society of Japan.
- Yamamoto, T., et al. (2016). The CCR4-NOT deadenylase: a key player of novel paradigm of generegulation. The 89th Annual Meeting of the Japanese Biochemical Society. Tohoku University.
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 Seminar:
6.1.1 Regulation of cell-fate decisions by MAPK signaling pathways and its failure in cancer
• Date: April 6, 2016
• Venue: OIST Campus Lab1
• Speaker: Dr. Mutsuhiro Takekawa
• Affiliation: Division of Cell Signaling and Molecular Medicine, Institute of Medical Science, The University of Tokyo
6.1.2 Screening of phosphorylation dependent protein-protein interaction inhibitors
• Date: April 13, 2016
• Venue: OIST Campus Lab1
• Speaker: Dr. Nobumoto Watanabe
• Affiliation: Bio-Active Compounds Discovery Research Unit, RIKEN Center for Sustainable Resource Science
6.1.3 Role of dsRNA pathways and miRNA-regulatory machinery in obesity
• Date: June 23, 2016
• Venue: OIST Campus Lab1
• Speaker: Dr. Takahisa Nakamura
• Affiliation: Divisions of Endocrinology and Developmental Biology, Cincinnati Children’s Hospital Medical Center
6.1.4 Translational capacity of a cell is determined during transcription elongation via the Ccr4-Not complex
• Date: September 26, 2016
• Venue: OIST Campus Lab1
• Speaker: Dr. Martine Collart
• Affiliation: Department of Microbiology and Molecular Medicine, University of Geneva
6.1.5 Small molecule inhibitors of cancer stem cells identified through target-based and cell based screening
• Date: Feb 1, 2017
• Venue: OIST Campus Lab1
• Speaker: Dr. Nobumoto Watanabe
• Affiliation: Bio-Active Compounds Discovery Research Unit, RIKEN Center for Sustainable Resource Science
6.1.6 DNA barcode technologies for high-throughput measurements of molecular and cellular dynamics
• Date: Feb 8, 2017
• Venue: OIST Campus Lab1
• Speaker: Dr. Nozomu Yachie
• Affiliation: Synthetic Biology Division, Research Center for Advanced Science and Technology, The University of Tokyo
6.1.7 Novel regulatory mechanisms of p53 action by androgen
• Date: Feb 15, 2017
• Venue: OIST Campus Lab1
• Speaker: Dr. Satoshi Inoue
• Affiliation: Department of Functional Biogerontology, Tokyo Metropolitan Institute of Gerontology
6.1.8 Therapeutic strategies targeting cancer stem cells
• Date: March 8, 2017
• Venue: OIST Campus Lab1
• Speaker: Dr. Hideyuki Saya
• Affiliation: Division of Gene Regulation, Institute of Advanced Medical Research (IAMR), Keio University School of Medicine
6.1.9 Single live-cell imaging of the non-canonical NF-kB pathway
• Date: March 22, 2017
• Venue: OIST Campus Lab1
• Speaker: Dr. Junichiro Inoue
• Affiliation: Department of Cancer Biology, Institute of Medical Science, The University of Tokyo
6.1.10 Identification of a novel p53 downstream pathway important in neuroendocrine tumor development
• Date: March 29, 2017
• Venue: OIST Campus Lab1
• Speaker: Dr. Rieko Ohki
• Affiliation: Laboratory of Fundamental Oncology, National Cancer Center Research Institute
6.2 Meeting:
6.2.1 CCR4-NOT Meeting
• Date: April 4-6, 2016
• Venue: OIST Campus Center Bld.
• Organizer: Keiji Kuba (Akita University), Tadashi Yamamoto (OIST)
• The number of Participants: 24
7. Other
Nothing to report.



