FY2018 Annual Report
Cell Signal Unit
Professor Tadashi Yamamoto

Abstract
The Cell Signal Unit studies molecular and cellular events that are relevant to and important for maintaining healthy life in response to environmental changes. The Unit explores the cause of various diseases that include cancer, neuronal disorder, immunological diseases, diabetes/obesity, and defects in development at the molecular level. To approach this issue, the Unit characterizes regulation of gene expression, particularly post-transcriptional regulation that includes regulation by microRNA, and long non-coding RNA and RNA binding proteins. The Unit also characterizes protein kinase-mediated cell signaling especially in the control of the brain function such as emotions, learning and memory.
1. Staff
- Dr. Patrick Stoney, Staff Scientist
- Dr. Ken Matsuura, Staff Scientist
- Dr. Akiko Yanagiya, Staff Scientist
- Dr. Akinori Takahashi, Postdoctoral Scholar
- Dr. Kristopher Paraiso Montrose, Postdoctoral Scholar
- Dr. Shou Soeda, Postdoctoral Scholar
- Dr. William Ashworth, Postdoctoral Scholar
- Dr. Olga Elisseeva, Visiting Researcher
- Ms. Saori Nishijima, Technical Staff
- Ms. Risa Ishida, Technical Staff
- Ms. Melissa Germany, Technical Staff
- Ms. Nao Ohmine, Technical Staff
- Ms. Atsuko Sato, Technical Staff
- Ms. Sandrine Anne Laure Burriel, Graduate Student
- Mr. Haytham Mohamed Aly Mohamed, Graduate Student
- Mr. Shohei Takaoka, Graduate Student
- Mr. Hemanta Sarmah, Graduate Student
- Ms. Dina Mostafa, Graduate Student
- Mr. Mohieldin Magdy Mahmoud Youssef, Graduate Student
- Mr. Hong Huat Hoh, Graduate Student
- Ms. Yuki Nakagawa, Research Unit Administrato
2. Collaborations
2.1 Physiology and Molecular Cellular Biology of the CCR4-NOT complex
- Description: Analyze the physiological and molecular biological roles of each component of the CCR4-NOT complex using gene-modified mice
- Type of collaboration: Joint research
- Researchers:
- I: Keiji Kuba, Department of Physiology, Graduate School of Medicine, Akita University
- II. Toru Suzuki, Laboratory of Immunogenetics, Riken Center for Integrative Medical Sciences
- III. Toshinobu Fujiwara, Laboratory of Biochemistry, Faculty of Pharmacy, Kinki University
- IV. Masahiro Morita and Nahum Sonenberg, Department of Biochemistry, McGill University.
2.2 Basic cancer research for prevention and treatment
- Description: Search for substances that contribute to the prevention and treatment of cancer from biological resources, and elucidate its mechanism of action
- Type of collaboration: Joint researches I and II
- Researchers:
- I. Representative director Kuniaki Nerome, Bioresource Laboratory LLC.
- II. Professor Shinya Ikematsu, National Institute of Technology, Okinawa College
2.3 Electrophysiology
- Description: Electrophysiological studies of gene modified mice with abnormal social behavior
- Type of collaboration: Joint research
- Researcher:
- Professor Toshiya Manabe, Institute of Medical Science, University of Tokyo
2.4 Mechanisms and physiological roles of CCR4-NOT complex-regulated gene expression in the brain
- Description: Investigation of mechanisms and physiological roles of CCR4-NOT complex-regulated gene expression in the brain
- Type of collaboration: Joint research
- Researcher:
- Team leader Masaru Tamura, RIKEN
2.5 Study of Cell-to-cell Interaction within Tumor Microenvironment Using An In-Vitro Three Dimensional Pancreatic Cancer Model
- Description:
- Establishing an in vitro 3D organoid culture for pancreatic ductal adenocarcinoma to study cell-cell interaction within tumor microenvironment
- Investigating the origin of cancer stem cells in pancreatic ductal adenocarcinoma by live cell imaging using tumor organoid
- Characterizing the effects of the interaction between stromal cells and cancer stem cells
- Identifying the targets for tumor-stroma crosstalk and cancer stem cells
- Type of collaboration: Joint research
- Researchers:
- Professor Masafumi Nakamura, Assistant Professor Kenoki Ohuchida, Kazuhiro Koikawa, Department of Surgery and Oncology, Graduate School of Medical Sciences, Kyushu University
2.6 Human immune responses and their modulation in cancer and autoimmunity
- Description: Human immune responses and their modulation in cancer and autoimmunity
- Type of collaboration: Joint research
- Researchers:
- Research Scientist Olga Elisseeva, Laboratory of Immunogenetics, Riken Center for Integrative Medical Sciences
3. Activities and Findings
3.1 The CCR4-NOT Deadenylase Complex Controls Atg7-dependent Cell Death and Heart Function
Shortening and removal of the polyadenylate [poly(A)] tail of mRNA, a process called deadenylation, is a key step in mRNA decay that is mediated through the CCR4-NOT (carbon catabolite repression 4-negative on TATA-less) complex. In our investigation of the regulation of mRNA deadenylation in the heart, we found that this complex was required to prevent cell death. Conditional deletion of the CCR4-NOT complex components Cnot1 or Cnot3 resulted in the formation of autophagic vacuoles and cardiomyocyte death, leading to lethal heart failure accompanied by long QT intervals. Cnot3 bound to and shortened the poly(A) tail of the mRNA encoding the key autophagy regulator Atg7. In Cnot3-depleted hearts, Atg7 expression was posttranscriptionally increased. Genetic ablation of Atg7, but not Atg5, increased survival and partially restored cardiac function of Cnot1 or Cnot3 knockout mice. We further showed that in Cnot3-depleted hearts, Atg7 interacted with p53 and modulated p53 activity to induce the expression of genes encoding cell death-promoting factors in cardiomyocytes, indicating that defects in deadenylation in the heart aberrantly activated Atg7 and p53 to promote cell death. Thus, mRNA deadenylation mediated by the CCR4-NOT complex is crucial to prevent Atg7-induced cell death and heart failure, suggesting a role for mRNA deadenylation in targeting autophagy genes to maintain normal cardiac homeostasis.
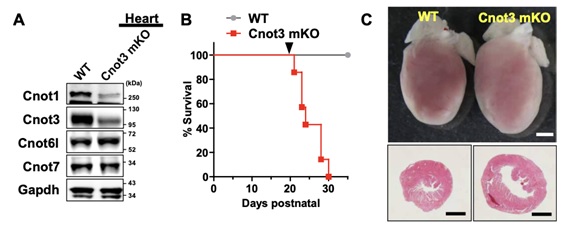
Figure 1: Severe heart failure by muscle-specific deletion of Cnot3 in mice. (A) Western blotting confirming suppression of Cnot3 as well as Cnot1 but not Cnot6l and Cnot7 proteins in the heart of Cnot3 muscle KO mice. (B) Postnatal survival curve for wild-type and Cnot3 muscle KO mice. (C) Macroscopic pictures and sections of hearts of wild-type and Cnot3 muscle KO mice.
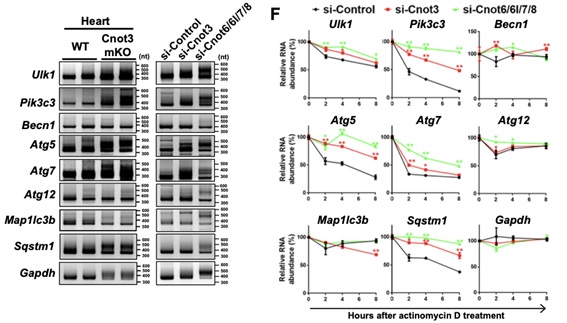
Figure 2. Poly(A) tail length and stability of autophagy factor-encoding mRNAs. (A) Poly(A) tail length measurement of autophagy factor-encoding mRNAs in hearts (left) and mouse cardiomyocytes in the presence and absence of Cnot3. (B) The stability of autophagy factor-encoding mRNAs in cardiomyocytes shown by actinomycin-chase experiments.
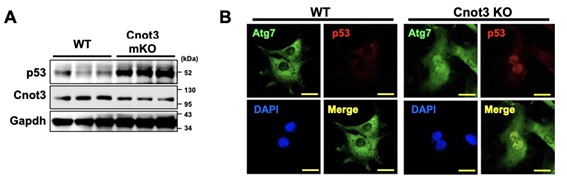
Figure 3. ATG7 regulates p53 activity to induce expression of cell death-associated genes under Cnot3 depletion. (A) Immuno-blotting showing elevated expression of p53 in Cnot3-depleted mouse hearts. (B) Immunocytochemistry showing co-localization of ATG7 with p53 in Cnot3 depleted MEFs.
3.2 Postnatal Liver Functional Maturation Requires Cnot Complex-Mediated Decay of mRNAs Encoding Cell Cycle and Immature Liver Genes
Liver development involves dramatic gene expression changes mediated by transcriptional and post-transcriptional control. Here, we show that the Cnot deadenylase complex plays a crucial role in liver functional maturation. The Cnot3 gene encodes an essential subunit of the Cnot complex. Mice lacking Cnot3 in liver have reduced body and liver masses, and they display anemia and severe liver damage. Histological analyses indicate that Cnot3-deficient (Cnot3-/- ) hepatocytes are irregular in size and morphology, resulting in formation of abnormal sinusoids. We observe hepatocyte death, increased abundance of mitotic and mononucleate hepatocytes, and inflammation. Cnot3-/- livers show increased expression of immune response-related, cell cycle-regulating and immature liver genes, while many genes relevant to liver functions, such as oxidation-reduction, lipid metabolism and mitochondrial function, decrease, indicating impaired liver functional maturation. Highly expressed mRNAs possess elongated poly(A) tails and are stabilized in Cnot3-/- livers, concomitant with an increase of the proteins they encode. In contrast, transcription of liver function-related mRNAs was lower in Cnot3-/- livers. We detect efficient suppression of Cnot3 protein postnatally, demonstrating the crucial contribution of mRNA decay to postnatal liver functional maturation.
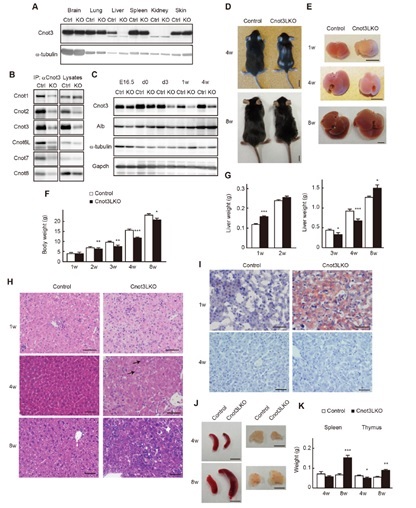
Figure 1. Liver-specific Cnot3 suppression results in decreased body mass and abnormal liver histology. (A) Cnot3 protein expression in tissues. (B) Expression of Cnot proteins in anti-cnot3 immuno-precipitates. (C) Cnot3 protein levels in developmental stages. (D and E) Appearance of whole body and liver. (F) Body weights. (G) Liver weights. (H) Liver H&E staining (I) Oil O-Red stained liver. (J) Appearance and (K) weights of the spleen and thymus.
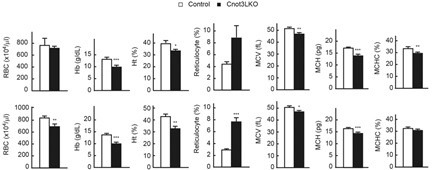
Figure 2 Liver specific Cnot3 depletion in mice causes ion-deficiency anemia. RBC, Red blood cells; Hb, hemoglobin; Ht, Hematocrit; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; Upper graphs, 4 weeks old mice; Lower graph 7-8 weeks old mice.
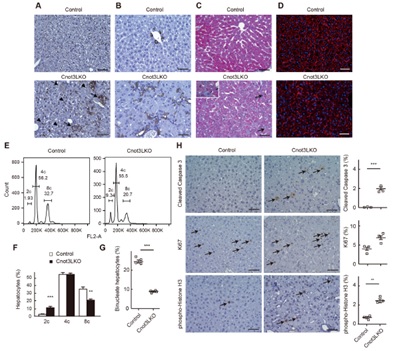
Figure 3. Increment of bile duct reaction, apoptotic cells, dividing cells, mononucleated cells in Cnot3-/- livers. (A-D) immunohistochemistry for F4/80 (A) and CK19 (B). Masson’s trichrome staining (C), and actin staining (D) of livers. (E,F) Identification of cells with diploid, tetraploid, and octaploid. (G) The percentage of binucleate hepatocytes. (H) Immunohistochemistry for cleaved caspase 3, Ki67, and phosphor-histone H3.
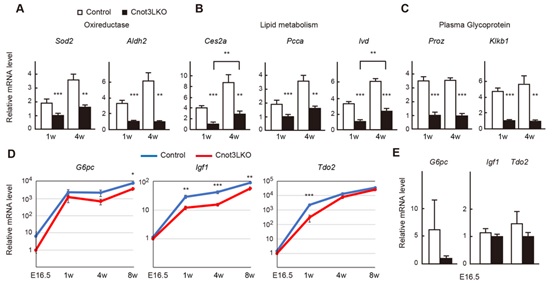
Figure 4. Genes involved in liver function display decreased expression in postnatal Cnot3-/- livers. (A) qPCR analysis
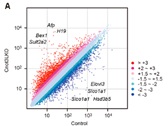
Figure 5. Scatter plot of mRNA expression in the control and Cnot3-depleted livers
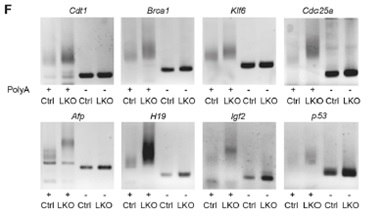
Figure 6. Comparison of Poly(A) tail length of mRNAs in the control and Cnot3-depleted livers.
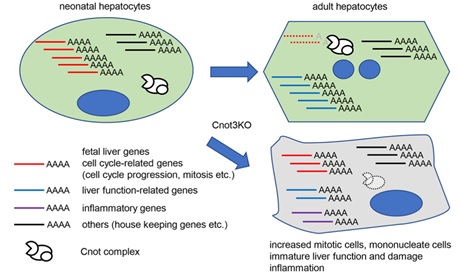
Figure 7 A schematic model of roles of Cnot3-mediated deadenylation in postnatal liver functional maturation.
4. Publications
4.1 Journals
- Morita M, Siddiqui N, Katsumura S, Rouya C, Larsson O, Nakashima T, Hekmatnejad B, Takahashi A, Kiyonari H, Zang M, St-Arnaud R, Oike Y, Giguère V, Topisirovic I, Okada-Hatakeyama M, Yamamoto T, Sonenberg N. Hepatic post-transcriptional network comprising of CCR4-NOT deadenylase and FGF21 maintains systemic metabolic homeostasis. Proc Nat Acad Sci USA 116:7973-7981, 2019
- Shirai Y, Mizutani A, Nishijima S, Horie M, Kikuguchi C, Elisseeva O, Yamamoto T. CNOT3 targets negative cell cycle regulators in non-small cell lung cancer development. Oncogene 38:2580-2594, 2019
- Suzuki T, Kikuguchi C, Nishijima S, Nagashima T, Takahashi A, Okada M, Yamamoto T. Postnatal liver functional maturation requires Cnot complex-mediated decay of mRNAs encoding cell cycle and immature liver genes. Development 146:dev168146, 2019
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- Yamamoto, T. Cdk1-mediated DIAPH1 phosphorylation maintains metaphase cortical tension and inactivates the spindle assembly checkpoint at anaphase. The 16th International Membrane Research Forum. OIST, Japan, 2019.03.18 (2019).
- Matsuura, K, Iwasaki, K, Negishi, R, Senda, T, Nakamura, T, Kawahara, S, Kirino, Y, Yamamoto, T, Manabe, T, Akiyama, T. SPAL1 interacts with the Neurabin family of proteins and regulates G Protein¬-Coupled Receptor signaling. The 5th Core-to-Core International Symposium "3D Lab¬Exchange Program", OIST, Japan, 2019.02.25 (2019)
- Ashworth, W, Stoney, P, Yamamoto, T. Investigating the roles of the CCR4-NOT deadenylase subunit Cnot7 in the brain. The 5th Core-to-Core International Symposium "3D Lab¬Exchange Program", OIST, Japan, 2019.02.25 (2019)
- Takaoka, S, Takahashi, A, Mohamed, Haytham M.A., Yamamoto, T. Mathematical analysis of the importance of mRNA decay in circadian rhythm. The 1st International symposium on interdisciplinary approaches to integrative understanding of biological signaling networks. Tokyo, Japan, 2019.02.01 (2019)
- Yanagiya, A, Mostafa, D, Yamamoto, T. Post-transcriptional regulation in insulin biosynthesis by the Ccr4-Not deadenylase complex in mouse pancreatic islets. The 20th Takeda Science Foundation Symposium on BioSciences, "RNA Neobiology", Osaka, Japan, 2019.02.01 (2019)
- Montrose, K, Kobayashi S, Manabe T, Yamamoto, T. Lmtk3 -KO mice exhibit a range of behavioural abnormalities and have impaired GluA1 trafficking. The University of Osaka and OIST Joint Symposium, Osaka, Japan, 2019.01.29 (2019)
- Mostafa, D, Takahashi, A, Yamamoto, T. Functional analysis of CCR4-NOT complex in pancreatic beta cells. University of Osaka and OIST Joint Symposium, Osaka, Japan, 2019.01.29 (2019)
- Takahashi, A, Takaoka, S, Yamaguchi, T, Mohamed, Haytham M.A., Kuba, K, Yamamoto, T. Deadenylation represses mRNA stability and transcription of cell death and inflammatory genes to suppress lethal hepatitis. The 41st Annual Meeting of the Molecular Biology Sciety of Japan, Yokohama, Japan, 2018.11.29 (2018)
- Soeda, S, Yamamoto, T. The role of maternal mRNA decay in mouse early development. The 41st Annual Meeting of the Molecular Biology Society of Japan, Yokohama, Japan, 2018.11.29 (2018)
- Nishijima, S, Suzuki, T, Hoshina, M, Hoshina, N, Takahashi, A, Yamamoto, T. Roles of Cnot complex-mediated mRNA decay in stress response. The 41st Annual Meeting of the Molecular Biology Society of Japan. Yokohama, Japan, 2018.11.28 (2018)
- Montrose, K, Kobayashi, S, Manabe, T, Yamamoto, T. Lmtk3-KO mice exhibit a range of behavioural abnormalities and have impaired GluA1 trafficking. Okinawa Clinical Simulation Center, Okinawa, Japan, 2018.11.22 (2018)
- Yamamoto, T. Physiology of mRNA poly(A) tail and CCR4-NOT. Institute of Genetics and Molecular Cellular Biology, Strasbourg, France, 2018.11.05 (2018)
- Ashworth, W, Davies, N, Bogle, D. Analysis of potential drug targets for non-alcoholic fatty liver disease computational se in a model. ICSB, Lyon, France, 2018.10.28 (2018)
- Sarmah, H, Ito, K, Kikuguchi, C, Yamamoto, T. Molecular and Physiological Function of Mammalian CNOT9. East Asia Joint Symposium, Chongqing, China, 2018.10.25 (2018)
- Yamamoto, T, Suzuki, T. Roles of mRNA decay machinery in our life. Riken Center for Integrative Medical Sciences, Yokohama, Japan, 2018.10.12 (2018)
- Vares, G. Addressing tumor resistance by combining CSC-targeting strategies and high-LET radiation therapy. The 77th Annual Meeting of the Japanese Cancer Association, Osaka, Japan, 2018.09.28 (2018)
- Burriel, S, Yamamoto, T. Lemur Tyrosine Kinase 1 and its Role in Lung Cancer: Computational and Functional Analysis. 1st Course on Computational Systems Biology of Cancer, Paris, France, 2018.09.27 (2018)
- Soeda, S, Yamamoto, T. The role of maternal mRNA decay in developmental ability in mammalian early embryos. RNA Frontier Meeting, Kanagawa, Japan, 2018.09.21 (2018)
- Takahashi, A, Mohamed, Haytham M.A., Takaoka, S, Asai, Y, Yamamoto, T. The importance of mRNA decay in circadian rhythm. Integrative understanding of biological signaling networks based on mathematical science - 2nd Young members workshop, Shiga, Japan, 2018.09.01 (2018)
- Stoney, Patrick N., Fang, Kristen A., Huntly, Rachael L., Yamamoto, T. Investigating the roles of the CCR4-NOT deadenylase subunit Cnot7 in the brain. FENS Forum of Neuroscience 2018, Berlin, Germany, 2018.07.11 (2018)
- Montrose, K, Kobayashi, S, Yamamoto, T. LMTK3 deficiency leads to impaired GluA1 trafficking and schizophrenia-like behaviour in mice. 11th FENS, Forum of Neuroscience 2018, Berlin, Germany, 2018.07.11 (2018)
- Takahashi, A, Takaoka, S, Mohamed, Haytham M.A., Yamaguchi, T, kuba, k, Yamamoto, T. Deadenylation-dependent mRNA decay regulates mRNA stability and transcription to maintain gene expression in liver. The 20th Annual Meeting of the RNA Society of Japan, Osaka, Japan, 2018.7.10 (2018)
- Youssef, Mohieldin M.M., Kiyama, Y, Hamada, H, Takahashi, A, Suzuki, T, Montrose, K, Stoney, Patrick N., Manabe, T, Yamamoto, T. Investigating physiological and stress-related Tob protein functions in the brain. FENS forum 2018, Berlin, Germany, 2018.07.08 (2018)
- Mostafa, D, Takahashi, A, Yanagiya, A, Yamamoto, T. CNOT3 maintains β-cell identity through repression of β-cell disallowed genes. The 6th CCR4-NOT Meeting, Wakayama, Japan, 2018.05.13 (2018)
- Soeda, S. RSK-MASTL pathway delays meiotic exit in mouse zygotes to ensure paternal. Joint Annual Meeting of 51st JSDB and 70th JSCB, Tokyo, Japan, 2018.06.07 (2018)
- Takahashi, A, Takaoka, S, Mohamed, Haytham M.A., Yamamoto, T. Deadenylation suppresses cell death and immune genes to maintain liver homeostasis. RNA 2018, The 23rd Annual Meeting of the RNA Society, UC Berkley, USA, 2018.06.01 (2018)
- Takahashi, A, Shohei T., Mohamed, Haytham A. M., Yamamoto, T. Deadenylation-dependent mRNA decay is required for liver homeostasis thorough suppression of cell death and inflammatory genes. Joint Annual Meeting of 51st JSDB and 70th JSCB, Tokyo, Japan, 2018.06.07 (2018)
- Takahashi, A, Yamamoto, T. The CCR4-NOT complex-mediated mRNA decay regulates gene expression and liver function. The 6th CCR4-NOT Meeting, Wakayama, Japan, 2018.05.13 (2018)
- Takahashi, A, Yamamoto, T. Deadenylation-dependent mRNA decay regulates gene expression in liver. The 10th Signal Network Meeting, Kobe, Japan, 2018.06.30 (2018)
- Yanagiya, A, Mostafa, D, Yamamoto, T. Post-transcriptional regulation in glucose-stimulated insulin biosynthesis by the Ccr4-Not deadenylase complex in mouse pancreatic islets. RNA 2018, The 23rd Annual Meeting of the RNA Society, UC Berkeley, USA, 2018.06.01 (2018)
5. Intellectual Property Rights and Other Specific Achievements
5.1 Intellectual Property RightsTitle: Anti-tumor agent
Patent Number: JP6443872B2
Inventors: Kuniaki Nerome, Tadashi Yamamoto, Taku Kureha, Reiko Nerome
Owner: The Institute of Biological Resources
Priority Date: Nov. 10, 2016
6. Meetings and Events
6.1 Seminar
6.1.1 Role of actin dynamics in reprogramming and fate determination of cells
- Date: April 4, 2018
- Venue: OIST Campus Lab1
- Speaker: Dr. Saya Hideyuki (Keio University School of Medicine)
6.1.2 Transcriptional control of human embryo genome activation
- Date: April 11, 2018
- Venue: OIST Campus Lab1
- Speaker: Dr. Juha Kere (Karolinska Institute)
6.1.3 Regulation of cell-fate decisions by stress-responsive p38/JNK MAPK signaling pathways
- Date: April 18, 2018
- Venue: OIST Campus Lab1
- Speaker: Dr. Mutsuhiro Takekawa (Institute of Medical Science, The University of Tokyo)
6.1.4 Toward the solution for neurodegenerative disorders with muscle atrophy: the theoretical consideration and two clinical examples of HMSN-p and myotonic dystrophy
- Date: August 7, 2018
- Venue: OIST Campus Lab1
- Speaker: Dr. Shugo Suwazono (National Hospital Organization Okinawa Hospital)
6.1.5 Imaging and single-cell analyses of sensory nerves in barrier-impaired, itchy skin
- Date: November 8, 2018
- Venue: OIST Campus Lab1
- Speaker: Dr. Takaharu Okada (RIKEN)
6.1.6 On the mechanism of biomolecular pattern self-organization for spatio-temporal control of cellular processes
- Date: November 18, 2018
- Venue: OIST Campus Lab1
- Speaker: Dr. Kiyoshi Mizuuchi (NIH, USA)
6.1.7 Polycomb in gene activation
- Date: December 3, 2018
- Venue: OIST Campus Lab1
- Speaker: Dr. Haruhiko Koseki (RIKEN)
6.1.8 Spatio-temporal dynamics of SAPKK regulates cell fate decisions under stress conditions
- Date: January 16, 2019
- Venue: OIST Campus Lab1
- Speaker: Dr. Mutsuhiro Takekawa (Institute of Medical Science, The University of Tokyo)
6.1.9 Telomere as the starting point of anticancer drug discovery
- Date: June 23, 2019
- Venue: OIST Campus Lab1
- Speaker: Dr. Seimiya Hiroyuki (Japanese Foundation for Cancer Research)
6.1.10 Translational Control of Cancer and Neurological Diseases via eIF4E
- Date: February 4, 2019
- Venue: OIST Campus Lab
- Speaker: Dr. Nahum Sonenberg (McGill University, Canada)
6.1.11 A novel therapeutic strategy for osteosarcoma by inducing terminal adipocyte differentiation in chemoresistant stemlike cells
- Date: February 6, 2019
- Venue: OIST Campus Lab1
- Speaker: Dr. Saya Hideyuki (Keio University School of Medicine)
6.1.12 Cutting-edge Basic and Translational Research in the Carbon-Ion Radiotherapy
- Date: February 8, 2019
- Venue: OIST Campus Lab1
- Speaker: Dr. Sei Sai (National Institute for Quantum and Radiological Science and Technology)
6.1.13 Healthy people with “clonal hematopoiesis” tend to develop a variety of adult diseases including leukemia, strokes and acute myocardial infarction
- Date: February 13, 2019
- Venue: OIST Campus Lab1
- Speaker: Dr. Kitamura Toshio (Institute of Medical Science, The University of Tokyo)
6.1.14 Small molecule inhibitors of cancer stem cells that target mitochondria
- Date: February 20, 2019
- Venue: OIST Campus Lab1
- Speaker: Dr. Watanabe Nobumoto (RIKEN)
6.1.15 Implications of ERBB2 amplification
- Date: February 27, 2019
- Venue: OIST Campus Lab1
- Speaker: Dr. Semba Kentaro (Waseda University)
6.1.16 Fast-in, Fast-out: Safe driving through mitotic transitions
- Date: March 6, 2019
- Venue: OIST Campus Lab1
- Speaker: Dr. Toru Hirota (Japanese Foundation for Cancer Research)
6.1.17 Identification of a novel p53 downstream pathway important in neuroendocrine tumor development
- Date: March 13, 2019
- Venue: OIST Campus Lab1
- Speaker: Dr. Rieko Ohki (National Cancer Center Research Institute)
6.1.18 Possible role of ADF/cofilin in fission yeast cytokinesis
- Date: March 20, 2019
- Venue: OIST Campus Lab1
- Speaker: Dr. Mabuchi Issei (Professor emeritus, The University of Tokyo)
6.1.19 Dynamic regulation of inhibition of T cell activation
- Date: March 27, 2019
- Venue: OIST Campus Lab1
- Speaker: Dr. Saito Takashi (RIKEN)
7. Other
Nothing to report.



