FY2022 Annual Report
Nucleic Acid Chemistry and Engineering Unit
Professor Yohei Yokobayashi
Abstract
FY2022 started with the publication of a collaborative work with the Biological Complexity Unit on genomic replication mechanism of E. coli. It was followed several articles from our Unit focusing on DNA and RNA chemistry and biology. We also focused on summarizing the results of our collaborative project with Astellas Pharma that concluded in FY2021. This work will be published early FY2023. On the personnel side, Takeshi Tabuchi successfully defended his thesis. Dr. Fukunaga moved on to a new faculty position at Tokyo Institute of Technlogy while we welcomed a new postdoc Dr. Bochen Zhu.
1. Staff
- Dr. Marius Baeken, Researcher
- Dr. Samuel Hauf, Researcher
- Dr. Narae Kim, Researcher
- Dr. Yoko Nomura, Science and Technology Associate
- Dr. Lara Sellés Vidal, Researcher
- Dr. Bochen Zhu, Researcher
- Rachapun (Gear) Rotrattanadumrong, Graduate Student
- Samira Gmür, Graduate Student
- Tomoya Noma, Graduate Student
- Nao Miyahira, Technical Staff
- Mayumi Suzuki, Technical Staff
- Yayoi Maehara, Technical Staff
- Hitomi Shinzato, Research Unit Administrator
(As of 3/31/2023)
2. Collaborations
2.1 Study on E. coli chromosome replication
- The main goal of this project is to understand how the speed of genome replication by DNA polymerase (replisomes) vary along the course of E. coli's replication cycle. Our unit performed bacterial culturing and genomic DNA isolation experiments.
- Researchers:
- Professor Simone Pigolotti, Biological Complexity Unit, OIST
- Dr. Deepak Bhat, Biological Complexity Unit, OIST
- Dr. Charles Plessy, Genomics and Regulatory Systems Unit, OIST
2.2 Collaboration with an industrial partner
- Undisclosed collaboration with an industrial partner
3. Activities and Findings
3.1 Experimental exploration of ribozyme neutral network
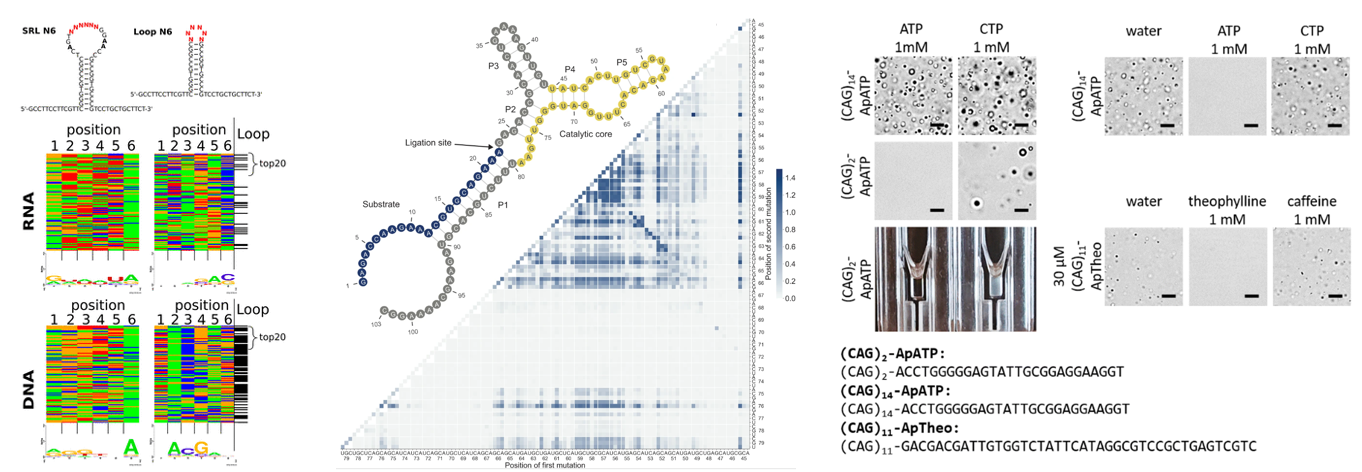
A neutral network connects all genotypes with equivalent phenotypes in a fitness landscape and plays an important role in the mutational robustness and evolvability of biomolecules. In contrast to earlier theoretical works, evidence of large neutral networks has been lacking in recent experimental studies of fitness landscapes. This suggests that evolution could be constrained globally. We demonstrated that a deep learning-guided evolutionary algorithm can efficiently identify neutral genotypes within the sequence space of an RNA ligase ribozyme. Furthermore, we measured the activities of all 216 variants connecting two active ribozymes that differ by 16 mutations and analyzed mutational interactions (epistasis) up to the 16th order. We discovered an extensive network of neutral paths linking the two genotypes and revealed that these paths might be predicted using only information from lower-order interactions. Our experimental evaluation of over 120,000 ribozyme sequences provides important empirical evidence that neutral networks can increase the accessibility and predictability of the fitness landscape.
3.2 Saporin substrate preference
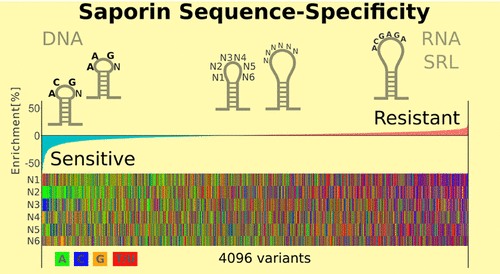
Ribosome-inactivating proteins (RIPs) are RNA:adenosine glycosidases that inactivate eukaryotic ribosomes by depurinating the sarcin-ricin loop (SRL) in 28S rRNA. The GAGA sequence at the top of the SRL or at the top of a hairpin loop is assumed to be their target motif. Saporin is a RIP widely used to develop immunotoxins for research and medical applications, but its sequence specificity has not been investigated. We combined the conventional aniline cleavage assay for depurinated nucleic acids with high-throughput sequencing to study sequence-specific depurination of oligonucleotides caused by saporin. Our data revealed the sequence preference of saporin for different substrates and show that the GAGA motif is not efficiently targeted by this protein, neither in RNA nor in DNA. Instead, a preference of saporin for certain hairpin DNAs was observed. The observed sequence-specific activity of saporin may be relevant to antiviral or apoptosis-inducing effects of RIPs. The developed method could also be useful for studying the sequence specificity of depurination by other RIPs or enzymes.
3.3 Chemical control of phase separation of DNA solution
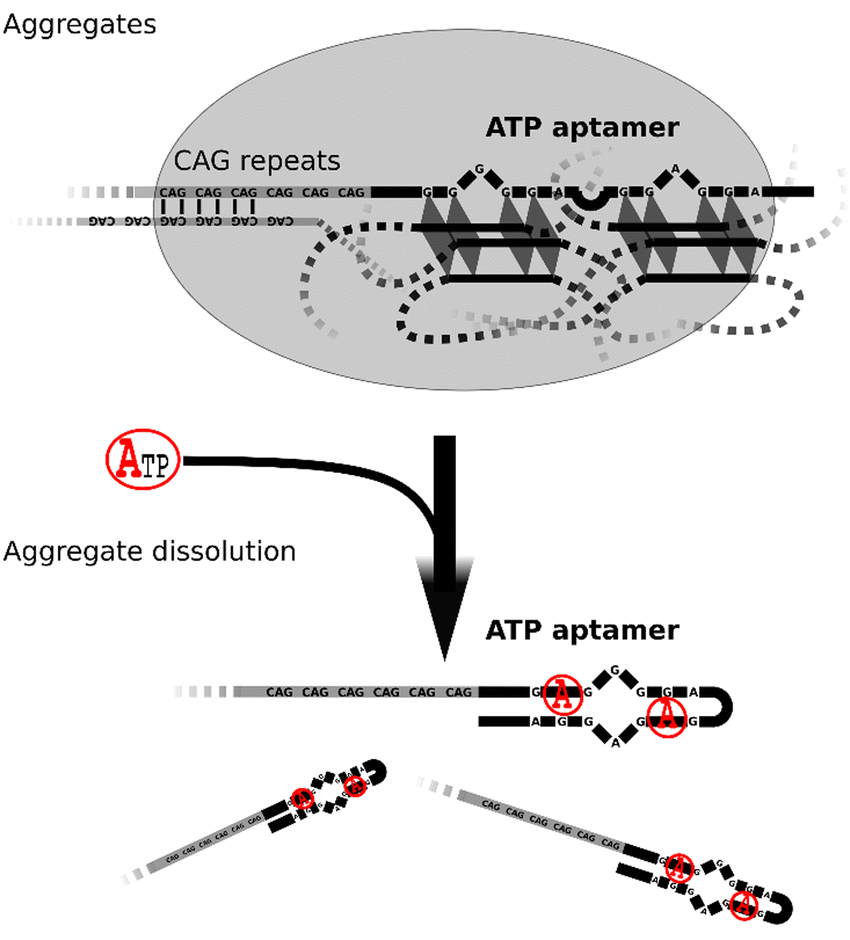
Phase separation is the process by which a solution of previously miscible chemical species becomes unmixed. Liquid–liquid phase separation (LLPS) has recently gained recognition as an important process in life, involved in regulatory processes, stress response, or certain diseases, among others. Besides its importance in natural phenomena, LLPS is also finding applications in synthetic biology. The process is usually driven by biopolymers such as proteins and/or nucleic acids forming dense intermolecular networks. Nucleic acids, especially RNA, are often involved in phase separation processes observed in cells. For example, RNA involved in repeat expansion disorders can form ribonucleoprotein bodies in the cellular nucleus. In the nucleolus, DNA, RNA, and proteins phase separate from the surrounding environment to regulate transcription. Stress granules are phase separated aggregates that increase fitness under stress conditions. How exactly phase separation occurs and how it is influenced by the sequence of the biopolymers are among the subjects of ongoing research. The process is assumed to be tightly controlled by factors such as the concentration and sequence of the phase separating species, posttranslational protein modifications (e.g., SUMOylation), temperature, and cation species. We designed a series of DNA sequences comprising a trinucleotide repeat segment and a small molecule-binding aptamer. Optimization of the DNA sequences and reaction conditions enabled chemical control of phase separation of DNA condensates. Our results demonstrate a new strategy to regulate biomolecular phase transition.
4. Publications
4.1 Journals
- Hauf S, Yokobayashi Y, Chemical control of phase separation in DNA solutions. Chem Commun 2023; 59: 3751-3754. (PubMed / Journal (Open Access))
- Rotrattanadumrong R, Yokobayashi Y, Experimental exploration of a ribozyme neutral network using evolutionary algorithm and deep learning. Nat Commun 2022; 13: 4847. (PubMed / Journal (Open Access)) OIST press release
- Hauf S, Rotrattanadumrong R, Yokobayashi Y, Analysis of the Sequence Preference of Saporin by Deep Sequencing. ACS Chem Biol 2022; 17: 2619-2630. (PubMed / Journal (Open Access))
- Baeken MW, Yokobayashi Y, Identification of an ERN1 target site within EGFP mRNA. J Cell Biochem 2022; 123: 1298-1305. (PubMed / Journal (Open Access))
- Bhat D, Hauf S., Plessy C, Yokobayashi Y, Pigolotti S. Speed variations of bacterial replisomes. eLife 2022; 11: e75884. (PubMed / Journal (Open Access))
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- Kim, N., Yokobayashi, Y. Application of RNA Viral Vector with Guanine Riboswitch to Primordial Germ Cell-Like Cell Induction, 2022 ISSCR Annual Meeting, hybrid (online/San Francisco, U.S.A.), June 15-18 (2022). (Poster)
- Rotrattanadumrong, R., Yokobayashi, Y. Experimental exploration of a ribozyme neutral network using evolutionary algorithm and deep learning, 30th Conference on Intelligent Systems for Molecular Biology, (Madison, U.S.A.), July 10-14 (2022). (Poster)
- Fukunaga, K., Yokobayashi, Y. 「試験管内共進化による分子パーツの機能改変」, 「超越分子システム」第2回領域会議, (Hokkaido, Japan), September 25-27, 2022. (Poster)
- Fukunaga, K., Yokobayashi, Y. Engineering RNAs and proteins by directed evolution, The 60th Annual Meeting of the Biophysical Society of Japan, hybrid (Hakodate, Japan), September 29 (2022). (Invited Talk)
- Tabuchi, T., Yokobayashi, Y. Chemical communication between microdroplets using cell-free riboswitches, The 49th International Symposium on Nucleic Acids Chemistry (ISNAC), (Tokyo, Japan), November 2-4 (2022). (Oral)
- Hauf, S., Rotrattanadumrong, R., Yokobayashi, Y. High-throughput analysis of a sequence-specific depurination enzyme, The 49th International Symposium on Nucleic Acids Chemistry (ISNAC), (Tokyo, Japan), November 2-4 (2022). (Poster)
- Kim, N., Yokobayashi, Y. NOVEL RNA VIRAL VECTORS CONTROL MYOGENIC DIFFERENTIATION IN MOUSE EMBRYONIC STEM CELLS BY A SMALL MOLECULE, ISSCR Boston International Symposium (Boston, U.S.A.), 17-19 November (2022). (Poster)
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 A small molecule targeting UGGAA pentanucleotide repeat responsible for spinocerebellar ataxia type 31
- Date: July 4, 2022
- Venue: OIST Campus Lab1
- Speaker: Prof. Tomonori Shibata (Osaka University)
6.2 HIV keeps on surprising us: a CRISPR-Cas cure adventure and a drug-resistance story
- Date: November 16, 2022
- Venue: OIST Campus Lab1
- Speaker: Prof. Ben Berkhout (University of Amsterdam)
7. Other
Degree awarded
- Takeshi Tabuchi, Ph.D. Sep 30, 2022.
Rotation students
- None
Research interns
- Theodor Röhrkasten (York University, Oct 2022 - Mar 2023)
Visiting faculty
- Prof. Ben Berkhout (University of Amsterdam, Nov 2022 - Jan 2023, JSPS Invitational Fellowship)
Outreach activities
- Theodor Röhrkasten, Dec 11, 2022, Science-Tech Festival (Naha, Okinawa), Making Möbius kaleidocycles.
- Theodor Röhrkasten, Jan 29, 2023, Industrial Tech Festival (Uruma, Okinawa), Making Möbius kaleidocycles.
External funding and support
- Yohei Yokobayashi: Undisclosed industry partner (continuing, PI), JSPS Grant-in-Aid for JSPS Fellows (continuing, PI/host researcher), JSPS Invitational Fellowships for Research in Japan (new, short-term, host), AMED/BINDS research support.
- Narae Kim: KAKENHI Young Resercher (new, PI) 22K15000
- Samuel Hauf: Walter Benjamin Fellowship (continuing)
- Keisuke Fukunaga: KAKENHI Kiban C (new, PI) 22K05322
- Lara Sellés Vidal: JSPS Postdoctoral Fellowship (continuing)
- Rachapun (Gear) Rotrattanadumrong: JSPS Predoctoral Fellowship (continuing)



