FY2021 Annual Report
Nucleic Acid Chemistry and Engineering Unit
Professor Yohei Yokobayashi
Abstract
FY2021 turned out to be the most productive year since the establishment of the unit at OIST, with a total of eight peer-reviewed articles published. We continued to develop new mammalian riboswitches and pursued their applications. Notably, we published several pieces of work on new topics, for example, macroscopic self-assembly and protein engineering of RNA-binding proteins. Furthermore, two new postdoctoral researchers overcame the COVID-19 related obstacles to join the unit, diversifying the repertoir of expertise represented by the unit members.
1. Staff
- Dr. Marius Baeken, Researcher
- Dr. Keisuke Fukunaga, Researcher
- Dr. Samuel Hauf, Researcher
- Dr. Narae Kim, Researcher
- Dr. Yoko Nomura, Science and Technology Associate
- Dr. Vyankat Sontakke, Researcher
- Dr. Lara Sellés Vidal, Researcher
- Takeshi Tabuchi, Graduate Student
- Rachapun (Gear) Rotrattanadumrong, Graduate Student
- Samira Gmür, Graduate Student
- Qiyi Qian, Rotation Student
- Nao Miyahira, Technical Staff
- Mayumi Suzuki, Technical Staff
- Hitomi Shinzato, Research Unit Administrator
- Yayoi Maehara, Research Assistant
(As of 3/31/2022)
2. Collaborations
2.1 Collaboration with an industrial partner (#1)
- Undisclosed collaboration with an industrial partner
2.2 Collaboration with an industrial partner (#2)
- Undisclosed collaboration with an industrial partner
2.3 Study on E. coli chromosome replication
- Biological Complexity Unit (Pigolotti Unit), OIST
3. Activities and Findings
3.1 Programmable macroscopic self-assembly driven by DNAs
DNAs exhibit ideal properties for self-assembly. They form highly predictable double-helix structures based on sequence complementarity which forms the chemical foundation of the life's ability to maintain and transfer genetic information. Researchers have long exploited the superb potential of DNAs to self-assemble in test tubes, a remarkable example being (2D and 3D) DNA origami. DNAs have also been used to assemble other materials such as polymer micorparticles and gold nanoparticles into ordered structures. We extended this DNA-programmed assembly to macroscale, something that we can see by our own eyes.
We prepared hydrogels with 1-2 mm edges and decorated their surfaces with short DNAs. When submersed in a buffer solution in a petri dish and shaken, these hydrogels assembled if and only if the DNA sequences were complementary to each other. We confirmed that sequence complementarity is required for self-assembly, and we could also disassemble the gels by adding a competing DNA strand.
This work demonstrates that DNAs can be used to program self-assembly of millimeter-sized objects. We plan to achieve more complex structures and dynamic regulation of macroscopic self-assembly using DNA. Conceptually, such programmable self-assembly at macroscopic scale may lead to applications in tissue engineering by encapsulating various cells in hydrogels. But it is also simply striking to observe supramolecular interactions operating at nanometer scale to drive self-assembly of visible objects.
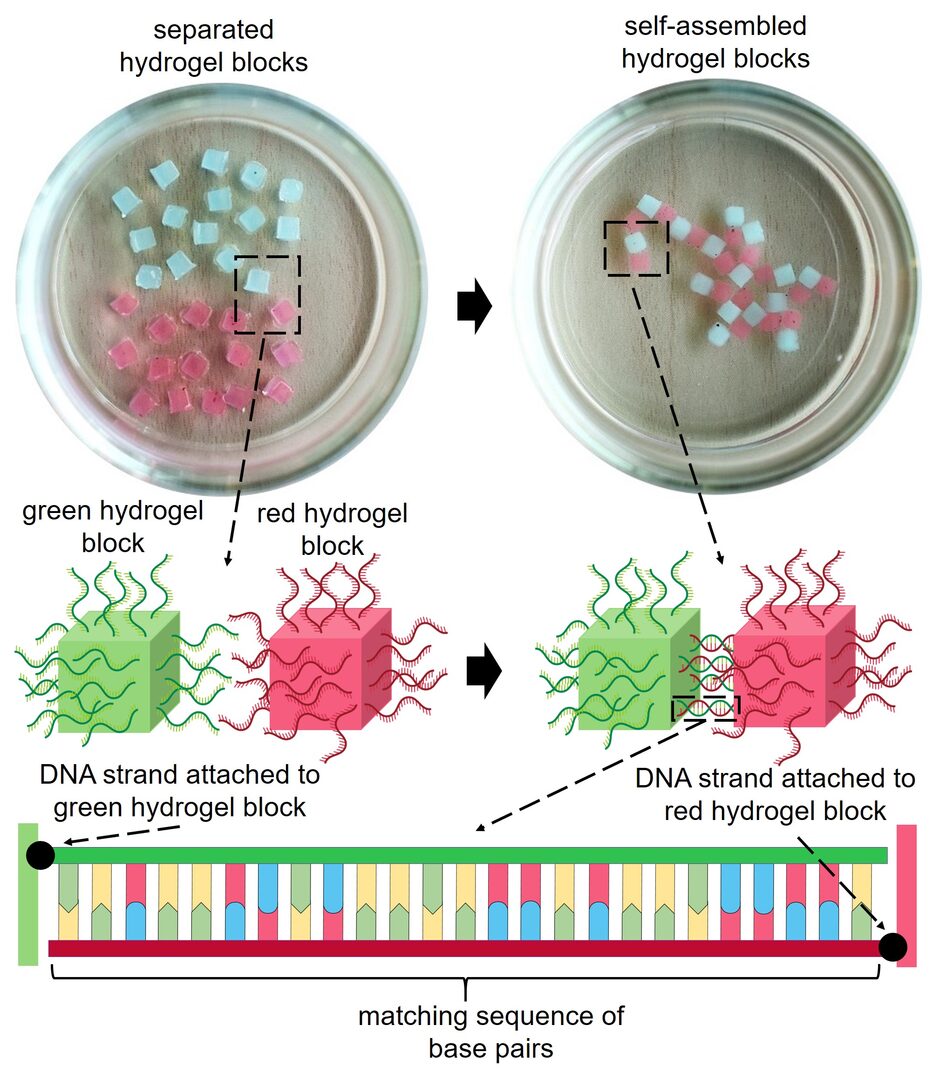
Figure 1. DNA-driven self-assembly of macroscopic hydrogels (image credit: OIST media section).
3.2 Directed evolution of RNA and RNA-binding protein pairs
(Adapted from https://doi.org/10.1093/nar/gkab527)
RNA-binding proteins (RBPs) and their RNA ligands play many critical roles in gene regulation and RNA processing in cells. They are also useful for various applications in cell biology and synthetic biology. However, re-engineering novel and orthogonal RNA–RBP pairs from natural components remains challenging while such synthetic RNA–RBP pairs could significantly expand the RNA–RBP toolbox for various applications. We developed a novel library-vs-library in vitro selection strategy based on Phage Display coupled with Systematic Evolution of Ligands by EXponential enrichment (PD-SELEX). Starting with pools of 1.1 × 1012 unique RNA sequences and 4.0 × 108 unique phage-displayed L7Ae-scaffold (LS) proteins, we selected RNA–RBP complexes through a two-step affinity purification process. After six rounds of library-vs-library selection, the selected RNAs and LS proteins were analyzed by next-generation sequencing (NGS). Further deconvolution of the enriched RNA and LS protein sequences revealed two synthetic and orthogonal RNA–RBP pairs that exhibit picomolar affinity and >4000-fold selectivity.
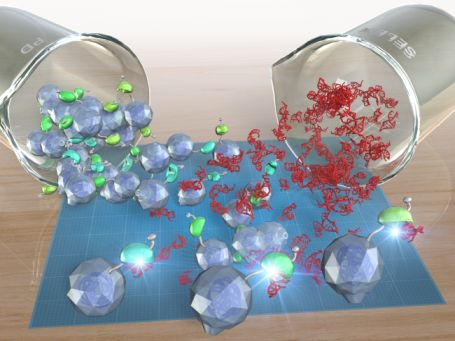
Figure 2. Artistic rendition of the PD-SELEX strategy.
3.3 Novel RNA viral vector that replicate in mouse ES cells
(Adapted from https://doi.org/10.1021/acssynbio.1c00214)
RNA viral vectors that replicate without DNA intermediates are attractive platforms for manipulation of cells for biomedical and veterinary applications because they have minimal risk of chromosomal integration. Vesicular stomatitis virus (VSV) vectors are among the most well-studied RNA viral vectors due to their low pathogenicity to humans and ability to express transgenes at high levels for weeks to months. However, their applications have been mostly limited to oncolytic and vaccine vectors due to their cytopathogenicity. We discovered two mutations in the VSV vector that synergistically confer improved stability in mouse embryonic stem cells (ESCs) with markedly lower cytopathic effects. We also demonstrated chemical regulation of transgene expression through embedded riboswitches. The ESCs infected with the mutant vector were shown to maintain pluripotency. This new vector sets the stage for precise regulation of gene expression in ESCs to produce a variety of differentiated cells without chromosomal alteration.
Figure 3. Mouse ES cells infected with the novel VSV mutant.
3.4 A synthetic ribozyme scaffold for mammalian riboswitches
(Adapted from https://doi.org/10.1021/acssynbio.1c00213)
A small molecule-responsive self-cleaving ribozyme (aptazyme) embedded in the untranslated region of an mRNA functions as a riboswitch that allows chemical regulation of gene expression in mammalian cells. Aptazymes are engineered by fusing a self-cleaving ribozyme with an RNA aptamer that recognizes a small molecule so that the ribozyme is either activated or inhibited in the presence of the small molecule. However, the variety of aptamers, ribozymes, and aptazyme design strategies suitable for mammalian riboswitch applications is still limited. This work focused on a new ribozyme scaffold for engineering aptazymes and riboswitches that function in mammalian cells. We investigated circularly permuted variants of the pistol ribozyme class (CPP) as a synthetic ribozyme scaffold for mammalian riboswitch applications. Through semirational design and high-throughput screening, we designed guanine and tetracycline activated riboswitches based on three distinct aptazyme architectures, resulting in riboswitches with ON/OFF ratios as high as 8.6. Our work adds CPP to the limited ribozyme scaffold toolbox for mammalian synthetic biology applications and highlights the opportunities in exploring ribozymes beyond natural motifs.
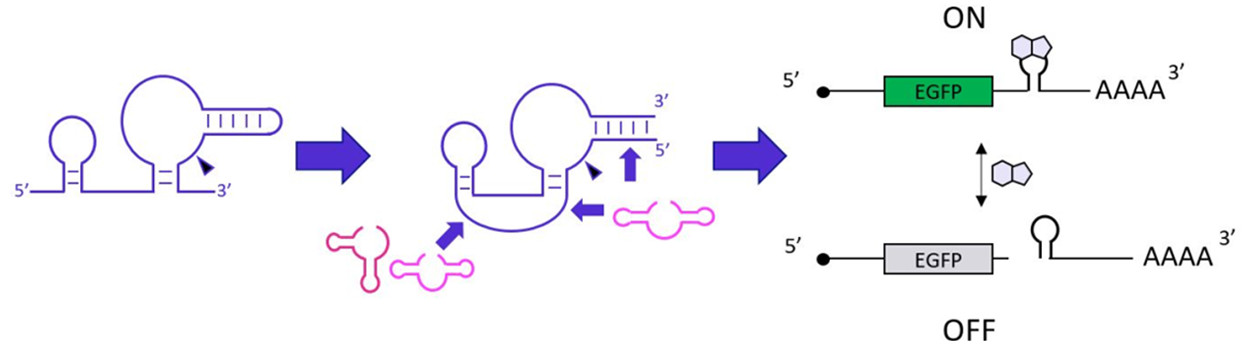
Figure 4. Schematic illustration of the CPP scaffold and its riboswitch derivatives.
3.5 High-throughput screening of cell-free riboswitches
(Adapted from https://doi.org/10.1093/nar/gkac152)
Cell-free systems that display complex functions without using living cells are emerging as new platforms to test our understanding of biological systems as well as for practical applications such as biosensors and biomanufacturing. Those that use cell-free protein synthesis (CFPS) systems to enable genetically programmed protein synthesis have relied on genetic regulatory components found or engineered in living cells. However, biological constraints such as cell permeability, metabolic stability, and toxicity of signaling molecules prevent development of cell-free devices using living cells even if cell-free systems are not subject to such constraints. Efforts to engineer regulatory components directly in CFPS systems thus far have been based on low-throughput experimental approaches, limiting the availability of basic components to build cell-free systems with diverse functions. Here, we report a high-throughput screening method to engineer cell-free riboswitches that respond to small molecules. Droplet-sorting of riboswitch variants in a CFPS system rapidly identified cell-free riboswitches that respond to compounds that are not amenable to bacterial screening methods. Finally, we used a histamine riboswitch to demonstrate chemical communication between cell-sized droplets.
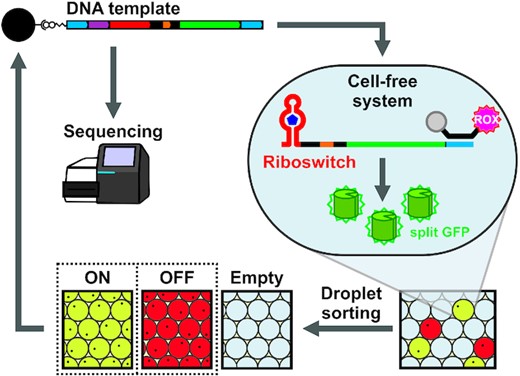
Figure 5. Schematic illustration of the high-throughput screening process.
4. Publications
4.1 Journals
- Tabuchi T, Yokobayashi Y. High-throughput screening of cell-free riboswitches by fluorescence-activated droplet sorting. Nucleic Acids Res 2022; DOI: 10.1093/nar/gkac152. (PubMed / Journal (Open Access))
- Sontakke VA, Yokobayashi Y. Programmable Macroscopic Self-Assembly of DNA-Decorated Hydrogels. J Am Chem Soc 2022; 144: 2149-2155. (PubMed / Journal (Open Access)) (Nature Resarch Highlight)
- Diaz Arenas C, Ardaševa A, Miller J, Mikheyev AS, Yokobayashi Y. Ribozyme Mutagenic Evolution: Mechanisms of Survival. Orig Life Evol Biosph 2022; DOI: 10.1007/s11084-021-09617-0. (PubMed / Journal)
- Kim N, Yokobayashi Y. Novel RNA Viral Vectors for Chemically Regulated Gene Expression in Embryonic Stem Cells. ACS Synth Biol 2021; 10: 2959-2967. (PubMed / Journal)
- Mustafina K, Nomura Y, Rotrattanadumrong R, Yokobayashi Y. Circularly-Permuted Pistol Ribozyme: A Synthetic Ribozyme Scaffold for Mammalian Riboswitches. ACS Synth Biol 2021; 10: 2040-2048. (PubMed / Journal)
- Tabuchi T, Yokobayashi Y. Cell-free riboswitches. RSC Chem Biol 2021; 2: 1430-1440. (PubMed / Journal (Open Access)) [Review]
- Fukunaga K, Yokobayashi Y. Directed evolution of orthogonal RNA-RBP pairs through library-vs-library in vitro selection. Nucleic Acids Res 2022; 50: 601-616. (Selected as a "Breakthrough Article". Cover picture.) (PubMed / Journal (Open Access))
- Nomura Y, Yokobayashi Y. Aptazyme-Based Riboswitches and Logic Gates in Mammalian Cells. Methods Mol Biol 2021; 2323: 213-220. (PubMed / Journal) [Method]
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- Yokobayashi, Y. Chemical Gene Regulation by Synthetic Riboswitches (人工リボスイッチによる化学的遺伝子発現制御), Japan Association for Chemical Innovation (JACI), Life Sciences Technical Division, Reactions Group Forum (新化学技術推進協会ライフサイエンス技術部会反応分科会勉強会), online, July 15 (2021). (Invited talk)
- Yokobayashi, Y. Engineering RNA Gene Switches, The University of Tokyo and OIST Joint Talk Series for Future Science, Season 4, online, July 28 (2021). (Invited talk)
- Yokobayashi, Y. Synthetic Mammalian Riboswitches, 2021 Cold Spring Harbor Asia Conference on Synthetic Biology, hybrid (online/Suzhou, China), October 27 (2021). (Invited talk)
- Yokobayashi, Y. High-throughput analysis of ribozyme sequence-function relationship, RIKEN BDR Symposium 2022 Emergence in Biological Systems: Challenges to Bridging Hierarchies, (online), March 1 (2022). (Invited Talk)
- Yokobayashi, Y. Engineering RNA Gene Switches (RNA遺伝子スイッチを創る), RNA Frontier Meeting 2021, (online), March 4 (2022). (Invited Talk)
- Fukunaga, K., Yokobayashi, Y. Directed evolution of orthogonal RNA–RBP pairs through library-vs-library in vitro selection, Webinar Universe 10 in FIBER - The Japan Society of Nucleic Acids Chemistry Forum for Young Researchers (日本核酸化学会若手フォーラム), online, August 5 (2021). (Research talk: Awarded "FIBER核酸化学若手講演賞")
- Fukunaga, K., Yokobayashi, Y. PD-SELEX法を用いたRNA-RBPのin vitro共進化, Summer School on the Society for the Study of the Origin and evolution of life - Japan (2021年生命の起原および進化学会「夏の学校」), online, September 2 (2021). (Research talk)
- Fukunaga, K., Yokobayashi, Y. ライブラリー vs. ライブラリーの試験管内選択を通した直交性RNA-RBPペアの発見, The 15th Symposium on Biorelevant Chemistry (第15回バイオ関連化学シンポジウム), online, September 9 (2021). (Research talk: Awarded "部会講演賞")
- Fukunaga, K., Yokobayashi, Y. PD-SELEX法を用いた直交性RNA-RBPペアの発見, 73rd SBJ Annual Meeting (第73回日本生物工学会大会), online, October 28 (2021). (Research talk)
- Fukunaga, K., Yokobayashi, Y. Directed evolution of orthogonal RNA-RBP pairs through library-vs-library in vitro selection, ISNAC2021, online, November 12 (2021). (Poster presentation)
- Fukunaga, K., Yokobayashi, Y. Directed evolution of orthogonal RNA-RBP pairs through library-vs-library in vitro selection, ICBS2021, online, November 12 (2021). (Poster presentation)
- Fukunaga, K., Yokobayashi, Y. Directed evolution of orthogonal RNA-RBP pairs through library-vs-library in vitro selection, RNA Frontier Meeting 2021, online, March 3 (2022). (Poster presentation)
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
Nothing to report
7. Other
Rotation students
- Maria Fernanda Bolanos Alejos (AY20, Term 3)
- Samira Gmür (AY20, Term 3)
- Tomoya Noma (AY21, Term 1)
External funding and support
- Yohei Yokobayashi: Undisclosed industry partner (continuing, PI), Undisclosed industry partner (new, PI), KAKENHI Kiban B 19H02855 (continuing, PI), JSPS Grant-in-Aid for JSPS Fellows (new, PI/host researcher)
- Narae Kim: KAKENHI Young Resercher (continuing, PI) 20K15669
- Samuel Hauf: Walter Benjamin Fellowship (continuing)
- Keisuke Fukunaga: KAKENHI Young Resercher (continuing, PI) 19K15701
- Lara Sellés Vidal: JSPS Postdoctoral Fellowship (new)
- Rachapun (Gear) Rotrattanadumrong: JSPS Predoctoral Fellowship (new)



