FY2016 Annual Report
Nucleic Acid Chemistry and Engineering Unit
Associate Professor Yohei Yokobayashi
Abstract
In FY2016, we significantly improved our deep sequencing based assay of ribozymes by demonstrating an exhaustive analysis of all single and double mutants of a self-cleaving ribozyme (>10,000 mutants). This technical milestone paves the way for more extensive and practical applications of our methodology to engineer synthetic riboswitches to control gene expression in mammalian cells. We also continued to push other efforts to design synthetic functional nucleic acids including bacterial riboswitches and in vitro evolution of ribozymes.
1. Staff
- Dr. Carolina Díaz Arenas, Researcher
- Dr. Mohammed Dwidar, Researcher
- Dr. Shungo Kobori, Researcher
- Dr. Yoko Nomura, Science and Technology Associate
- Dr. Kei Takahashi, Researcher
- Dr. Dhamodharan Venugopal, Researcher
- Kamila Mustafina, Graduate Student
- Takeshi Tabuchi, Rotation Student
- Crystal-Leigh Clitheroe, Technical Staff
- Hitomi Shinzato, Administrative Assistant
(As of 3/31/2017)
2. Collaborations
2.1 In vivo 3D Imaging Using Bioluminescent Gene Reporters and MRI
- We have been collaborating with two groups at University of California, Davis to develop a novel MRI probes to monitor gene expression in living animals.
- Type of collaboration: Joint research
- Researchers:
- Professor Angelique Louie, University of California, Davis
- Professor Jared Shaw, University of California, Davis
3. Activities and Findings
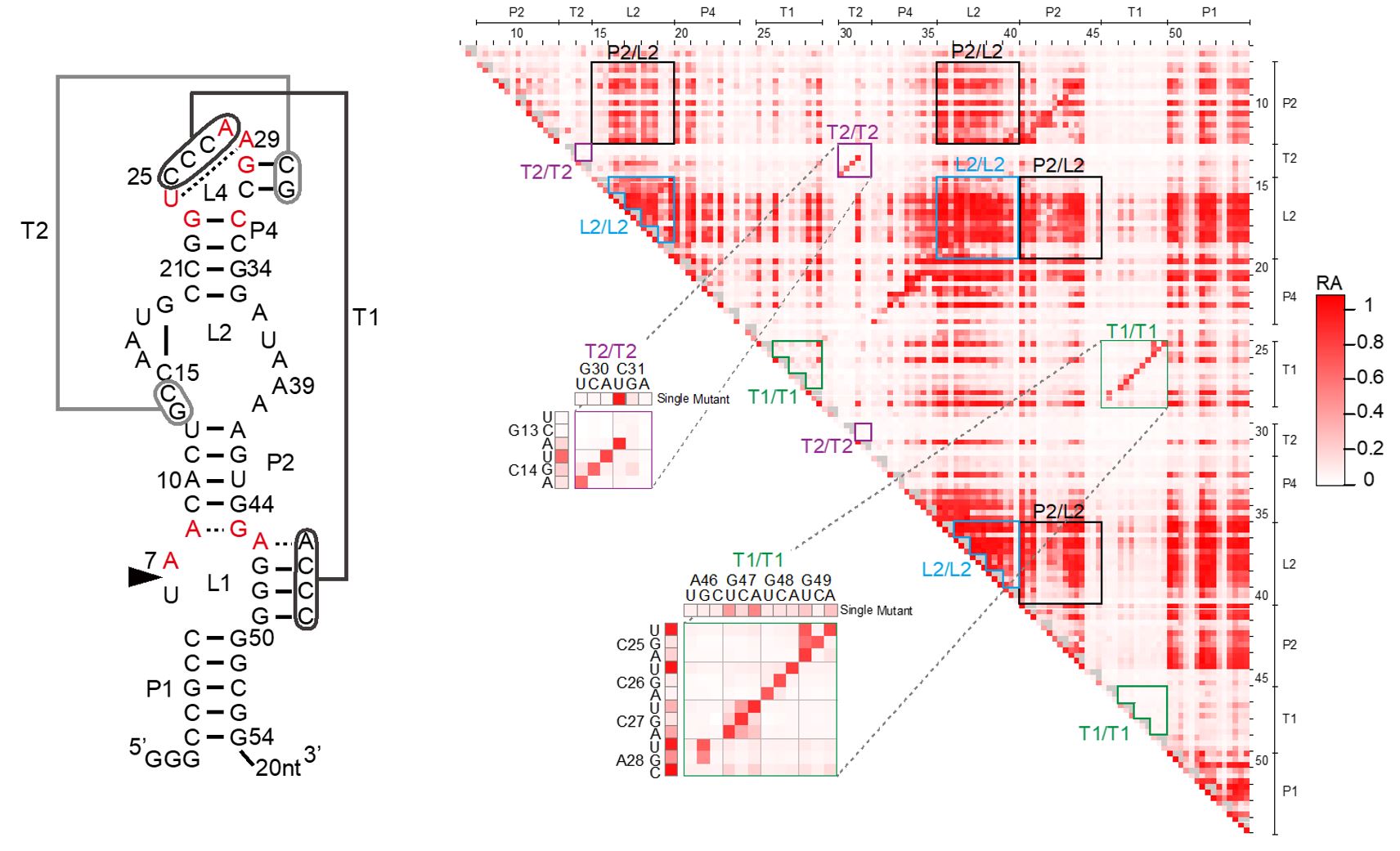
3.1 High-Throughput Characterization of Ribozymes by Deep Sequencing
Ribozymes are RNA molecules with catalytic activity. Natural ribozymes with RNA cleavage and splicing activities have been discovered and they are believed to play important roles in gene regulation as well as early evolution of life. Moreover, we and others have successfully engineered ribozymes to control gene expression in living cells, with potential future applications in biotechnology and medicine. However, characterizing ribozymes involves tedious preparation of many ribozyme mutants or variants and their activity must be measured individually. A method that allows efficient and quantitative characterization of many ribozyme variants would greatly deepen our understanding of ribozyme functions and our ability to engineer better ribozymes. To that end, we developed and reported a high-throughput ribozyme assay strategy based on deep sequencing in FY15.
Osa-1-4 twister ribozyme
This year, we extended our methodology to exhaustively analyze >10,000 mutants of a small self-cleaving ribozyme found in the rice genome (Figure 1). The mutants include all possible single and double mutants of the 54-nucleotide sequence. The results provided, for the first time, quantitative effects of all single and double mutations of a ribozyme, revealing its functionally critical nucleotides and mutational robustness and sensitivity.
3.2 Synthetic Bacterial Riboswitches
Riboswitches are chemically responsive gene regulatory elements that use RNA for both molecular recogniation (sensing) and gene regulation. Synthetic riboswitches have promising applications in biotechnology and medicine by enabling cells to respond to arbitrary chemical cues to control any genes of interest. In FY2016, we continued our efforts to develop riboswitches that respond to a small molecule inflammatory signal for potential probiotics applications. Specifically, we successfully isolated an RNA aptamer for the target small molecule and performed chemical analyses (e.g. binding asssay).
We also developed riboswitches that function in the predatory bacterium Bdellovibrio bacteriovorus which is known to predate a wide spectrum of gram-negative bacteria, including pathogenic strains. Data are being collected for publications in the near future.
3.3 In Vitro Evolution of Ribozymes
Ribozymes serve as an attractive platform for asking some basic questions in evolution. By combining an in vitro transcription of DNA into RNA by RNA polymerase, and a reverse transcription of RNA into DNA by reverse transcriptase, one can "evolve" a ribozyme in a test tube and study how mutations affect the evolution of the ribozyme populations. We are specifically studying how the mutation rate (controlled by Mn2+ concentration) of the ribozyme affects the distribution of the ribozyme mutants in the evolving populations.
3.4 Mammalian Riboswitches
4. Publications
4.1 Journals
- Kobori S, Yokobayashi Y. High-throughput Mutational Analysis of a Twister Ribozyme. Angew Chem Int Ed 2016; 55: 10354-10357.
- Yokobayashi Y. RNA Devices That Control Gene Expression. Seibutsu-kogaku Kaishi 2016; 94: 190-193 (invited review in Japanese). 横林 洋平, 「RNAによる遺伝子発現制御デバイス」, 生物工学会誌 2016; 94: 190-193.
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- Yokobayashi Y, Kobori S. Advances in high-throughput ribozyme assay based on deep sequencing, The 43rd International Symposium on Nucleic Acid Chemistry, Kumamoto, Japan, Sep 27-29 (2016). Oral presentation
- Takahashi K, Kobori S, Yokobayashi Y. Engineering allosteric pistol ribozymes by deep sequencing, The 43rd International Symposium on Nucleic Acid Chemistry, Kumamoto, Japan, Sep 27-29 (2016). Poster
- Dwidar M, Yokobayashi Y. Controlling gene expression in a predatory bacterium using synthetic riboswitches, The 43rd International Symposium on Nucleic Acid Chemistry, Kumamoto, Japan, Sep 27-29 (2016). Poster
- Kobori S, Yokobayashi Y. リボザイムの網羅的解析を可能にするハイスループットアッセイの開発 (Development of high-throughput assay for comprehensive mutational analysis of ribozymes), The 39th Annual Meeting of the Molecular Biology Society of Japan, Yokohama, Japan, Nov 30 - Dec 2 (2016). Poster
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 Synthetic biology for magnetic nanoparticles production
- Date: June 24, 2016
- Venue: OIST Campus Lab 1
- Speaker: Dr. Maiko Furubayashi (Massachusetts Institute of Technology)
6.2 Evolutionary Design of biosynthetic pathways and regulatory networks
- Date: November 18, 2016
- Venue: OIST Campus Center Building
- Speaker: Prof. Daisuke Umeno (Chiba University)
7. Other
Nothing to report.



