FY2015 Annual Report
Nucleic Acid Chemistry and Engineering Unit
Associate Professor Yohei Yokobayashi

Abstract
Nucleic Acid Chemistry and Engineering Unit was physically established at OIST in July 2015. In FY2015, the Unit recruited six researchers and one technician. With researchers from various fields including microbiology, virology, biochemistry, synthetic chemistry, and biotechnology, we initiated multiple projects with the common goal to study and engineer nucleic acids for applications in chemistry and biology.
1. Staff
- Dr. Carolina Díaz Arenas, Researcher
- Dr. Mohammed Dwidar, Researcher
- Dr. Shungo Kobori, Researcher
- Dr. Yoko Nomura, Science and Technology Associate
- Dr. Kei Takahashi, Researcher
- Dr. Dhamodharan Venugopal, Researcher
- Crystal-Leigh Clitheroe, Technical Staff
- Hitomi Shinzato, Administrative Assistant
(As of 3/31/2016)
2. Collaborations
2.1 In Vivo 3D Imaging Using Bioluminescent Gene Reporters and MRI
- We have been collaborating with two groups at University of California, Davis to develop a novel MRI probes to monitor gene expression in living animals.
- Type of collaboration: Joint research
- Researchers:
- Professor Angelique Louie, University of California, Davis
- Professor Jared Shaw, University of California, Davis
3. Activities and Findings
3.1 High-Throughput Characterization Ribozymes by Deep Sequencing
Ribozymes are RNA molecules with catalytic activity. Natural ribozymes with RNA cleavage and splicing activities have been discovered and they are believed to play important roles in gene regulation as well as early evolution of life. Moreover, we and others have successfully engineered ribozymes to control gene expression in living cells, with potential future applications in biotechnology and medicine. However, characterizing ribozymes involves tedious preparation of many ribozyme mutants or variants and their activity must be measured individually. A method that allows efficient and quantitative characterization of many ribozyme variants would greatly deepen our understanding of ribozyme functions and our ability to engineer better ribozymes. To that end, we developed a high-throughput ribozyme assay strategy based on deep sequencing. As depicted in Figure 1, a pool of ribozyme mutants are synthesized by in vitro transcription from a degenerate DNA template. The ribozyme mutants are then reverse transcribed and converted into DNA templates for high-throughput sequencing. The sequencing data are analyzed to evaluate catalytic activity of every mutant present in the ribozyme pool. We demonstrated the efficiency of the method by evaluating the activity of approximately 1,500 ribozyme variants in a single experiment. Some of the ribozymes were further tested for their ability to control gene expression in living mammalian cells.
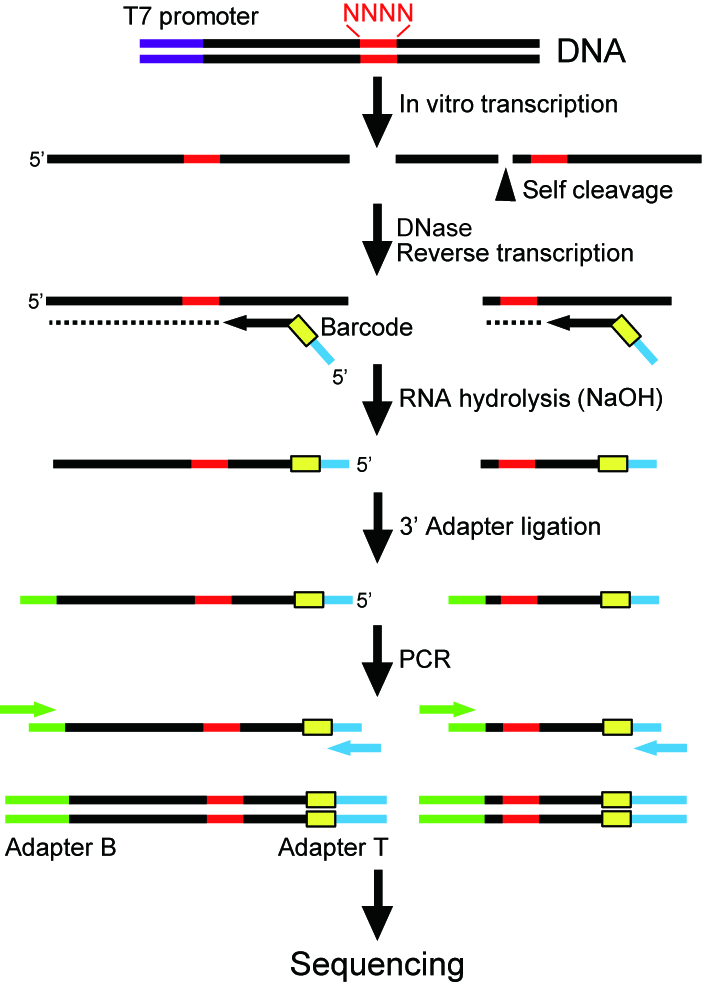
ribozyme assay.
3.2 Synthetic Bacterial Riboswitches
Riboswitches are chemically responsive gene regulatory elements that use RNA for both molecular recogniation (sensing) and gene regulation. Synthetic riboswitches have promising applications in biotechnology and medicine by enabling cells to respond to arbitrary chemical cues to control any genes of interest. In FY2015, we have started our efforts to 1) develop riboswitches that respond to a small molecule inflammatory signal for potential probiotics applications, and 2) develop riboswitches that function in the predatory bacterium Bdellovibrio bacteriovorus which is known to predate a wide spectrum of gram-negative bacteria, including pathogenic strains.
3.3 In Vitro Evolution of Ribozymes
Ribozymes serve as an attractive platform for asking some basic questions in evolution. By combining an in vitro transcription of DNA into RNA by RNA polymerase, and a reverse transcription of RNA into DNA by reverse transcriptase, one can "evolve" a ribozyme in a test tube and study how mutations affect the evolution of the ribozyme populations. We are specifically studying how the mutation rate (controlled by Mn2+ concentration) of the ribozyme affects the distribution of the ribozyme mutants in the evolving populations.
3.4 Artificial Nucleic Acids
4. Publications
4.1 Journals
- Nomura Y, Yokobayashi Y. Aptazyme-Based Riboswitches and Logic Gates in Mammalian Cells.Methods Mol Biol 2015; 1316: 141-148.
- Kobori S, Nomura Y, Miu A, Yokobayashi Y. High-throughput assay and engineering of self-cleaving ribozymes by sequencing. Nucleic Acids Res 2015; 43: e85.
- Akter F, Yokobayashi Y. RNA Signal Amplifier Circuit with Integrated Fluorescence Output. ACS Synth Biol 2015; 4: 655-658.
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- Kobori S, Nomura Y, Miu A, Yokobayashi Y. Parallel assay of self-cleaving ribozymes by high-throughput sequencing, The 42nd International Symposium on Nucleic Acid Chemistry, Himeji, Japan, Sep 23-25 (2015). Poster
- Yokobayashi Y. DNA and RNA Aptamers as Molecular Sensors, OIST Mini Symposium "Current Trends and Emerging Technologies in Nanofluidics and Nanofabrications", Oct 30-Nov 1 (2015). Invited talk
- Yokobayashi Y. RNAによる遺伝子発現制御デバイス作成のための手法(Methods for Developing RNA-Based Gene Regulatory Devices), 「細胞を創る」研究会8.0 (Japanese Society for Cell Synthesis Research Meeting), Osaka, Japan, Nov 12-13 (2015). Invited talk in Japanese
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 Synthetic biology for bioalcohol production
- Date: Dec 14, 2015
- Venue: OIST Campus Lab3
- Speaker: Dr. Taizo Hanai (Kyushu University)



