FY2017 Annual Report
Computational Neuroscience Unit
Professor Erik De Schutter

Abstract
We use computational, data-driven methods to study how neurons and microcircuits in the brain operate. We are interested in how fundamental properties, such as a neuron’s morphology and its excitability, interact with one another during common neural functions like information processing or learning. Most of our models concern the cerebellum as this brain structure has a relatively simple anatomy and the physiology of its main neurons has been studied extensively, allowing for detailed modeling at many different levels of complexity.
1. Staff
Molecular modeling
- Iain Hepburn, Technical Staff
- Andrew Gallimore, Researcher
- Criseida Zamora, Researcher
Cellular modeling
- Sungho Hong, Group Leader
- Yunliang Zang, Researcher
Network modeling
- Sergio Verduzco, Researcher
- Peter Bratby, Researcher
- Mykola Medvidov, Staff Scientist
Software development
- Weiliang Chen, Researcher
- Guido Klingbeil, Researcher
Visiting Researcher
- Akira Takashima (till May 2017)
- Yoshiyuki Kubota
- Jihwan Myung (till November 2017)
Research Interns
- Anna Valyaeva, Research Intern (July - October 2017)
- Ines Wichert, Research Intern (October - December 2017)
- Samyukta Chillara, Research Intern (October - January 2018)
- Merey Ramazanova, Research Intern (from January 2018)
Rotation Students
- Prasanna Vijayaraghavan, Student (Term 1)
- Wataru Ohata, Student (Term 1)
- Sarah Yukie Nagasawa, Student (Term 2)
- Thomas Burns (Term 3)
Research Unit Administrator
- Sachie Matsuoka (tlill January 2018)
- Keiko Hirano (till March 2018)
2. Collaborations
- Theme: Cerebellar physiology, multiple themes
- Type of collaboration: Scientific collaboration and graduate program
- Researchers:
- Professor M. Giugliano, University of Antwerp, Belgium
- Professor D. Snyders, University of Antwerp, Belgium
- Theme: Spiking activity of monkey cerebellar neurons
- Type of collaboration: Scientific collaboration
- Researchers:
- Professor H.P. Thier, University of Tübingen, Germany
- A. Ignashchenkova, University of Tübingen, Germany
- Dr. M. Junker, University of Tübingen, Germany
- A. Schmigdlin, University of Tübingen, Germany
- Theme: Human Brain Project: simulator development
- Type of collaboration: Scientific collaboration
- Researchers:
- Prof. F. Schürmann, École Polytechnique Fédérale de Lausanne, Switzerland
- F. Delalondre, École Polytechnique Fédérale de Lausanne, Switzerland
- Theme: Molecular identification of cerebellar signaling pathways and cerebellar optogenetics
- Type of collaboration: Scientific collaboration
- Researchers:
- Professor K. Tanaka, Korea Institute for Science and Technology (KIST), Korea
- Theme: Cerebellar molecular layer interneurons
- Type of collaboration: Joint research
- Researchers:
- Professor Alain Marty, Université Paris 5 René Descartes, France
- Theme: In vivo cerebellar activity
- Type of collaboration: Scientific collaboration
- Researchers:
- Professor C. De Zeeuw, Erasmus Medical Center, Rotterdam, The Netherlands
- Professor L.W.J. Bosman, Erasmus Medical Center, Rotterdam, The Netherlands
- Dr. M. Negrello, Erasmus Medical Center, Rotterdam, The Netherlands
- Theme: Purkinje cell morphology and physiology, modeling
- Type of collaboration: Scientific collaboration
- Researchers:
- Professor M. Häusser, University College London, United Kingdom
- Professor A. Roth, University College London, United Kingdom
- Dr. S. Dieudonné, Ecole Normale Supérieure, Paris, France
- Theme: Circadadian rhythm generation
- Type of collaboration: Scientific collaboration
- Researchers:
- Professor J. Myung, Taipei Medical University, Taiwan
3. Activities and Findings
3.1 Neuroinformatics standards
Updated standard file format for neuronal morphologies
Several efficient procedures exist to digitally trace neuronal structure from light microscopy, and mature community resources have emerged to store, share, and analyze these datasets based on the SWC file format (Figure 1, Cannon et al. 1998). In contrast, the quantification of intracellular distributions and morphological dynamics was not yet standardized. The SWC file format is static and inadequate for subcellular characterizations. In collaboration with Prof. G. Ascoli (George Mason University, USA) we extended the SWC file format to represent multichannel information and to capture morphological dynamics. This extension adds extra columns to the classic SWC format shown in Figure 1d and is fully back-compatible with popular visualization and modeling tools.
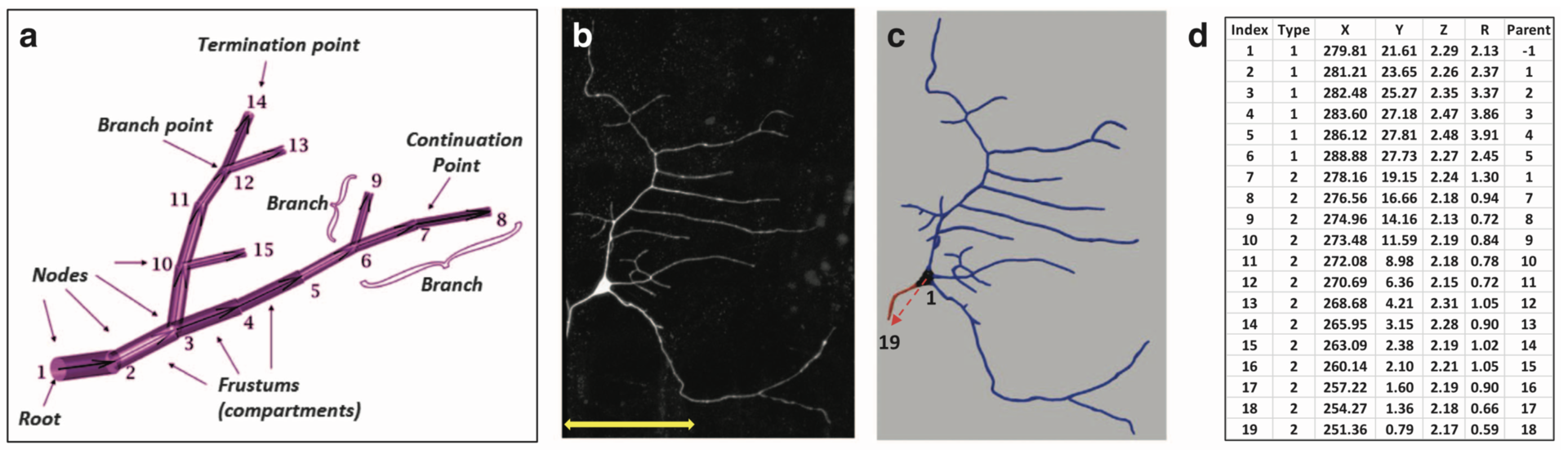
Figure 1: Examples of basic SWC reconstructions. (a) Representation of neural structure as a tree, along with graphical definitions of node, compartment, branch, branch point, continuation point, terminal point, and root node. (b) Image stack, (c) reconstruction, and (d) excerpt of swc file of Class I da neuron from the Drosophila larva. The SWC nodes marked with the red arrow in c are shown in d. The first six compartments described are soma (Type 1), followed by an axon (Type 2) coming off from the soma and going up to node 19. Dendritic nodes (Type 3) are not shown in d. (from Nandi et al. 2017)
In the resulting publication (Nandi et al. 2017) an application to combined multichannel/time-varying reconstruction data of developing neurons in live Drosophila larvae is shown. The files describe digitally tracing fluorescently labeled cytoskeletal components along with overall dendritic morphology as they changed over time.
3.2 Molecular mechanisms of synaptic plasticity
New model of bidirectional plasticity at parallel fiber to Purkinje cell synapses
Long-term depression (LTD) and long-term potentiation (LTP) in the cerebellum are important for motor learning. However, the signaling mechanisms controlling whether LTD or LTP is induced in response to synaptic stimulation remain obscure. We developed a unified model of LTD and LTP at the cerebellar parallel fiber-Purkinje cell synapse (Gallimore et al., 2018; model is summarized in Figure 2). It is comprised of 531 species, 358 reaction parameters, and 1,120 individual reactions, making it the most complex model of synaptic learning to date.
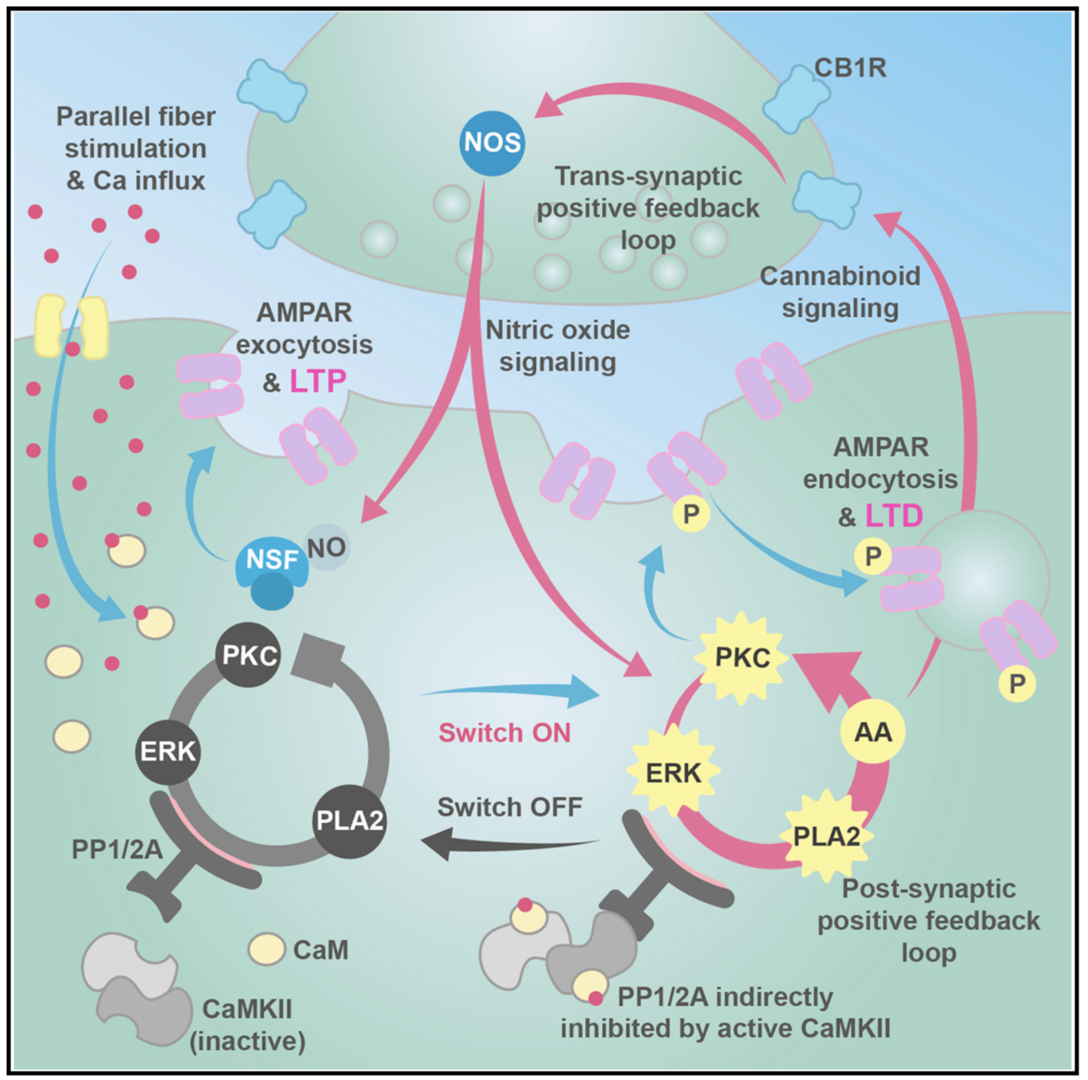
Figure 2: Unified model of LTP and LTD at the parallel fiber to Purkinje cell synapse. The system is activated by calcium entry which can activate cytosolic phospholipase A2 (cPLA2) leading to the production of cannabinoid 2-arachidonoylglycerol (2-AG) and presynaptic nitric oxide (NO) production, causing N-ethylmaleimide-sensitive factor (NSF)-dependent insertion of AMPA receptors into the postsynaptic membrane and LTP. Alternatively, larger calcium transients activate Ca2+/calmodulin-dependent protein kinase II (CaMKII) and cause LTD. In addition to the signaling pathways we also simulate a simple receptor trafficking model (from Gallimore et al. 2018)
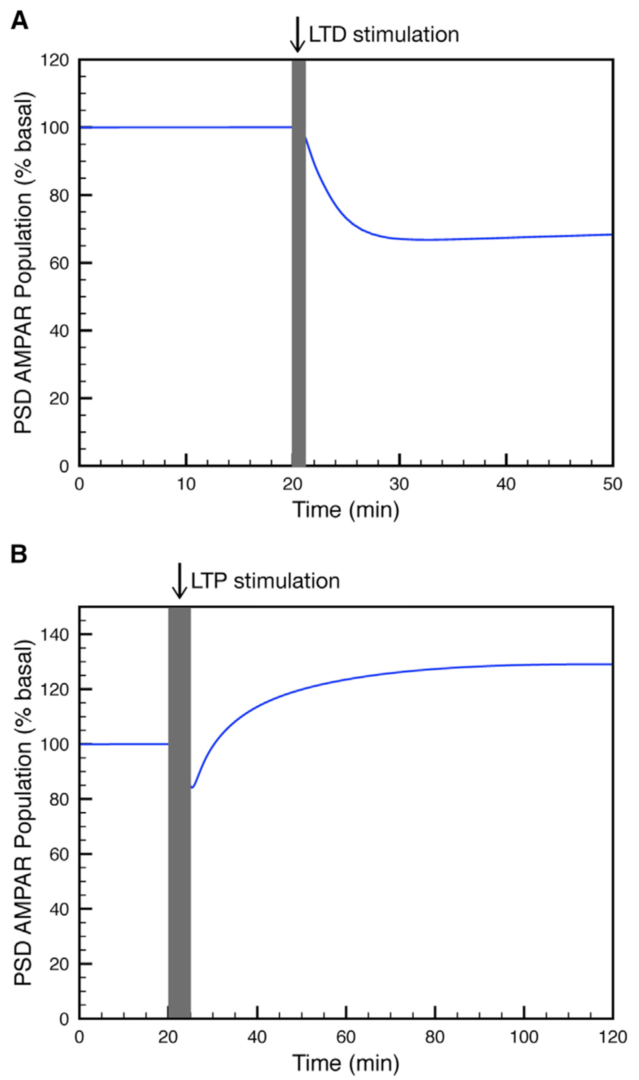
Figure 3: Simulation of (A) LTP or (B) LTD at the parallel fiber to Purkinje cell synapse. (from Gallimore et al. 2018)
Using this model we determined the coordinated pre- and postsynaptic signaling that determines the direction of plasticity. LTP is the default response to parallel fiber stimulation above a well-defined frequency threshold (Figure 3A). However, if the calcium signal surpasses the threshold for CaMKII activation, then an ultrasensitive ‘‘on switch’’ activates an extracellular signal-regulated kinase (ERK)-based positive feedback loop that triggers LTD instead (Figure 3B). This is possible because CaMKII activation leads to the suppression of protein phosphatase 2A (PP2A) which at basal conditions suppresses activation of several members of the feedback loop (Figure 4).
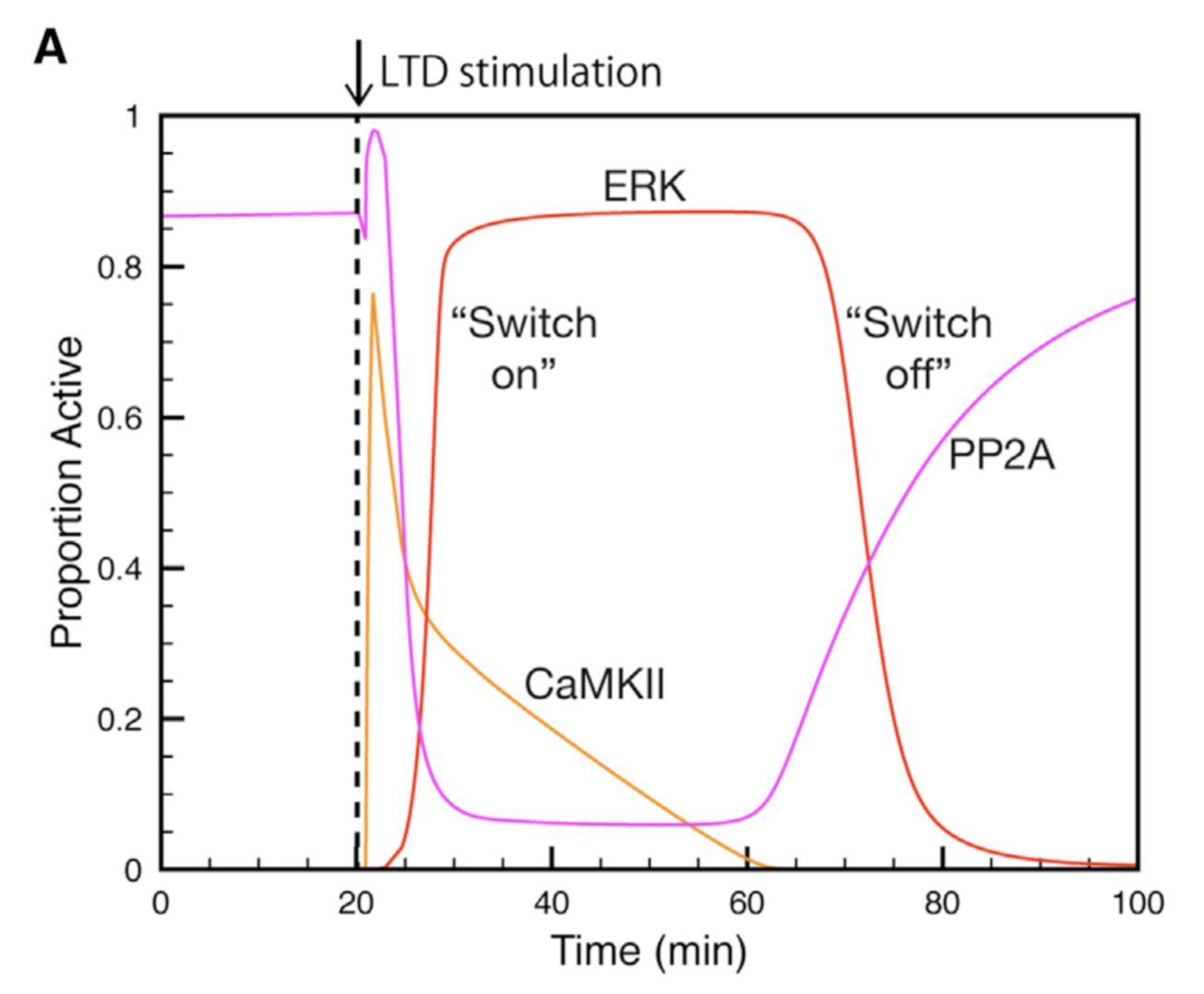
Figure 4: Activation and deactivation of the ERK-based feedack loop. Synaptic stimulation activates CaMKII which deactivates PP2A, allowing ERK to be activated. When CaMKII activity declines PP2A will shut ERK acitivity down (from Gallimore et al. 2018).
The postsynaptic ERK-based feedback loop is sustained by another, transsynaptic, feedback loop, activated by presynaptic cannabinoid receptors, that maintains nitric oxide production throughout LTD induction (Figure 2). When full depression is achieved, an automatic ‘‘off switch’’ inactivates the feedback loops, returning the network to its basal state and demarcating the end of the early phase of LTD (Figure 4).
3.3 Information processing in the olivocerebellar system
Firing dynamics in a large network model of the cerebellar granular layer
The cerebellar granular layer, which mainly consists of granule and Golgi cells, is the first stage of the cerebellar cortex and processes spatiotemporal information transmitted by mossy fiber inputs with a wide variety of firing patterns. To study its dynamics at multiple time scales in response to inputs approximating real spatiotemporal patterns, we constructed a large-scale 3D network model of the granular layer (Figure 5) comprising almost a million neurons and including signficant parallel fiber lengths (Sudhakar et al. 2017).
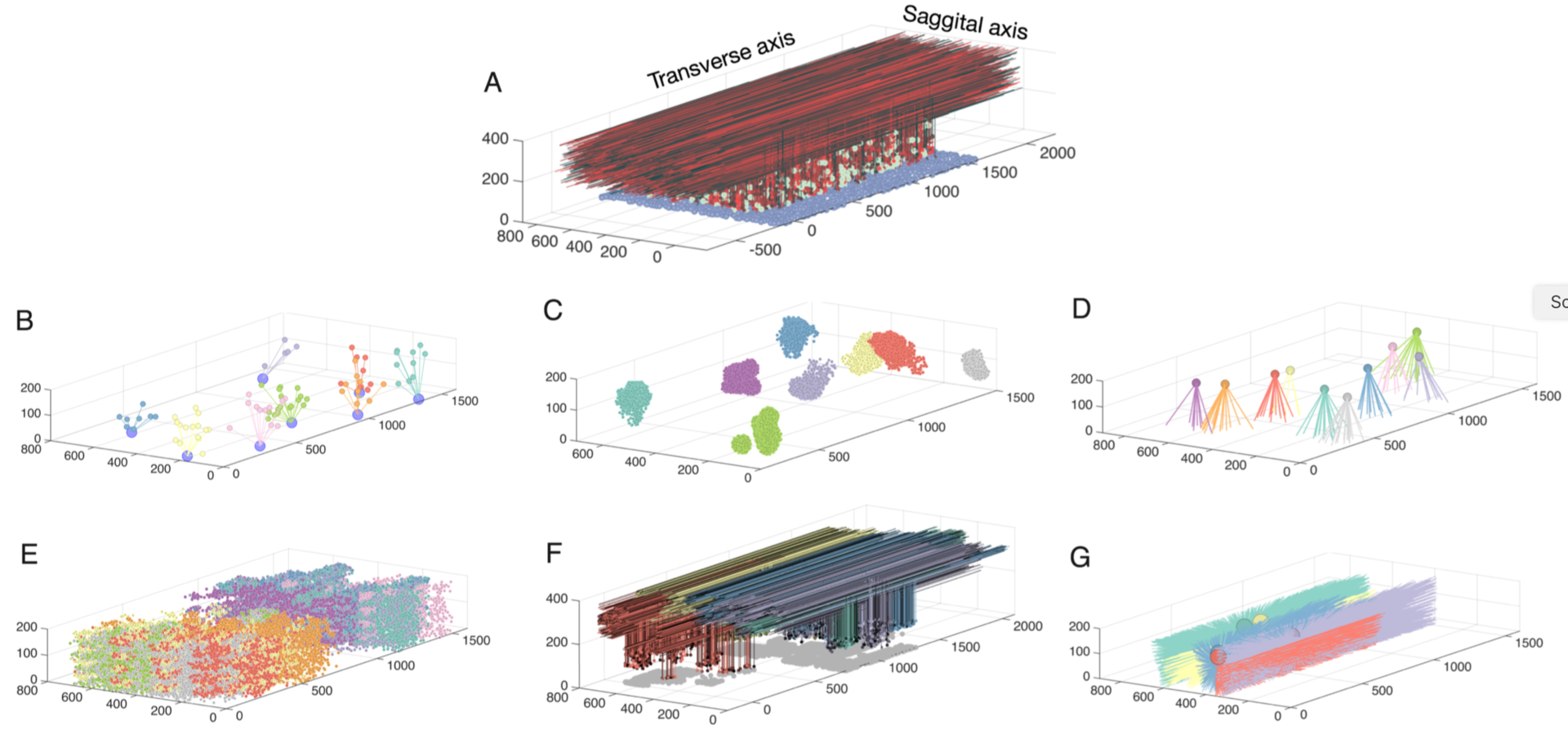
Figure 5: Network connectivity for all populations in the granule layer model. A: The completely assembled network showing 10% of all granule cells and parallel fibers (red shades), along with all Golgi cells (green) and all mossy fibers locations (blue). The following panels always show subsets of model elements to demonstrate their specific connectivity. B: Example of 7 mossy fibers diverging onto Golgi cells. C: The granule cells contacted by 8 different mossy fibers through rosettes (color coded by mossy fiber identity). D: Mossy fibers converging onto different Golgi cells (one color per Golgi cell). E.: granule cells inhibited by different Golgi cells (each color represents one Golgi cell). F: Ascending axons and parallel fibers relating to 5 different mossy fibers (each color represents one mossy fiber) G: Parallel fibers from granule cells, which converge onto different Golgi cells (each color represents one Golgi cell) (from Sudhakar et al. 2017).
Patterned mossy fiber activity induced rhythmic Golgi cell activity that is synchronized by shared parallel fiber input and by gap junctions (Figure 6). This led to long distance synchrony of Golgi cells along the transverse axis, powerfully regulating granule cell firing by imposing inhibition during a specific time window.
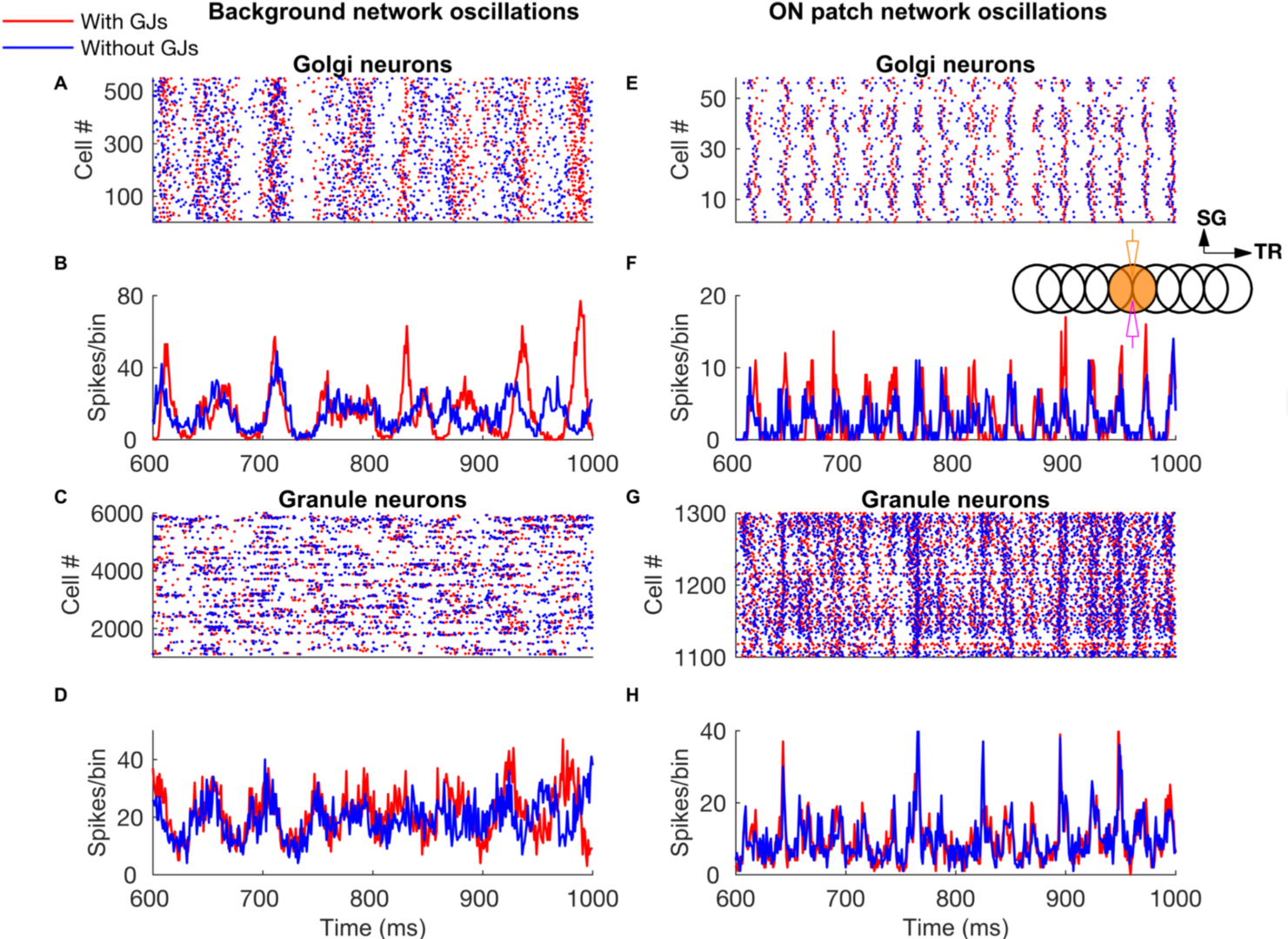
Figure 6: Network-wide oscillations with mossy fiber inputs. These oscillations are mostly driven by long-distance common parallel fiber input to the Golgi cells (blue traces, without Golgi cell gap junctions), though at low firing rates (panels A-D) the gap junctions also clearly contribute by making the oscillations more prominent (red traces). Local, patchy mossy fiber input increases oscillation frequencies and neural firing rates (E-H). A-D: Golgi cell and granule cell activity in the presence of 5 Hz background mossy fiber input. A and B represent Golgi cell population raster plot and population spike timing histogram, respectively. C and D are the same type of plots as A and B for the granule cell population. E-H: Golgi cell and granule cell activity when mossy fibers within a patch (a red circle in Fig 1) are activated at 60 Hz in addition to the 5 Hz mossy fiber activation of the rest of the network. E, F: Golgi cell raster plot and population spike timing histogram. G, H: the same plots for the granule cells. The inset represents stimulation and recording configuration used for E-H. An orange and empty region represents the stimulated (ON) and unstimulated (OFF) patch, respectively. Electrodes represent stimulation (orange) and recording (magenta), respectively. TR: Transverse. SG: Sagittal (from Sudhakar et al. 2017).
The essential network mechanisms, including tunable Golgi cell oscillations, on-beam inhibition and NMDA receptors causing first winner keeps winning of granule cells (Figure 7), illustrate how fundamental properties of the granule layer operate in tandem to produce (1) well timed and spatially bound output, (2) a wide dynamic range of granule cell firing and (3) transient and coherent gating oscillations.
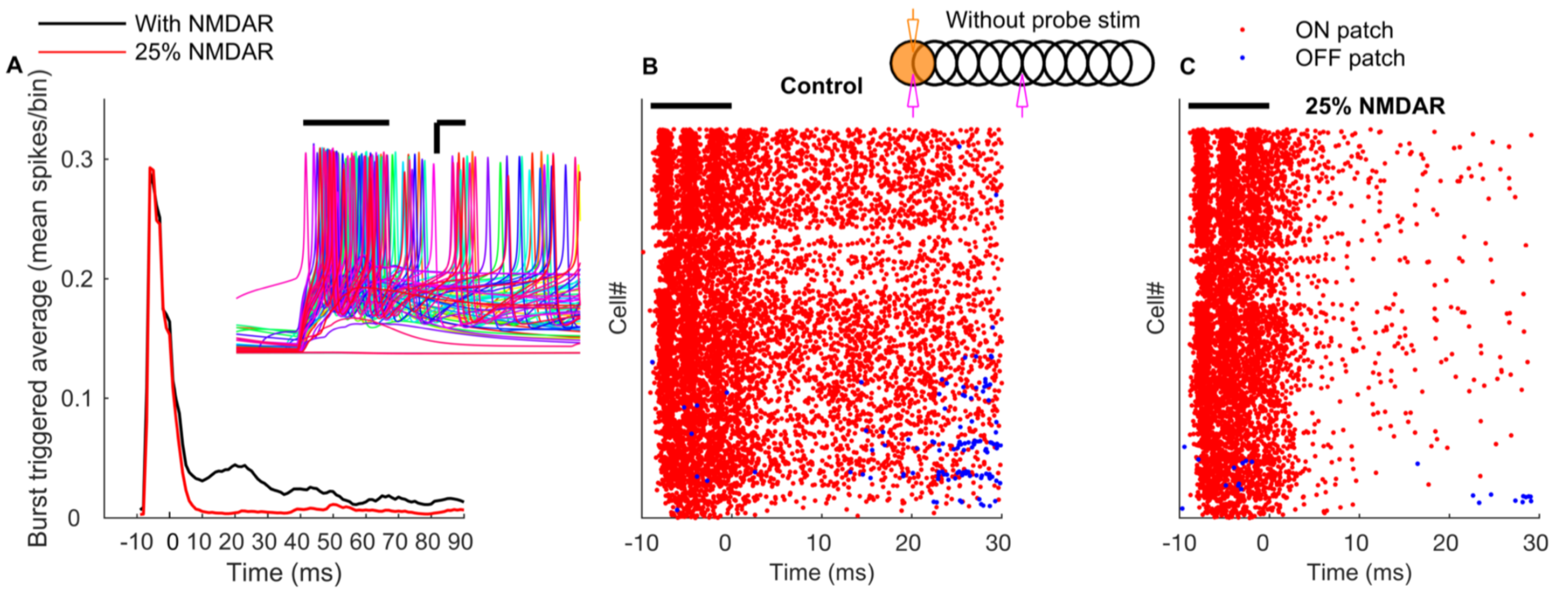
Figure 7: NMDA receptors mediate long-term gain increase in granule cells. A: Burst triggered average firing rate of the ON patch granule cells with normal or reduced conductance of NMDA receptors demonstrates a sustained increas in granule cell excitability due to activation of NMDA currents. Time zero corresponds to the end of the mossy fiber burst. Inset represents membrane potential traces of representative ON patch granule cells in response to bursting mossy fiber input. Scale bar: x-axis = 3 ms, y-axis = 15 mV. B: Raster plot of firing of granule cells in ON (red) and OFF patch (blue) in the absence of probe input. C: Same as B, with conductance of NMDA receptors reduced by 75%.
These results substantially enrich our understanding of granule cell layer processing, which seems to promote spatial group selection of granule cell activity as a function of timing of mossy fiber input.
Coupling between oscillators in circadian rhythm generation
Mammalian circadian clocks have a hierarchical organization, governed by the suprachiasmatic nucleus in the hypothalamus. The brain itself contains multiple loci that maintain autonomous circadian rhythmicity, but the contribution of clocks outsied of the suprachiasmatic nucleus to this hierarchy remains unclear.
We examined circadian oscillations of clock gene expression in various brain loci and discovered that in mouse, robust, higher amplitude, relatively faster oscillations occur in the choroid plexus compared to the suprachiasmatic nucleus (Myung et al. 2018). Our computational analysis and modeling showed that the choroid plexus achieves these properties by synchronization of “twist” circadian oscillators via gap-junctional connections. Using an in vitro tissue coculture model and in vivo targeted deletion of the Bmal1 gene to silence the choroid plexus circadian clock, we demonstrated that the choroid plexus clock adjusts the suprachiasmatic nucleus clock likely via circulation of cerebrospinal fluid, thus finely tuning behavioral circadian rhythms.
4. Publications
4.1 Journals
1. De Schutter, E.: Deep Learning and Computational Neuroscience. Neuroinformatics 16, 1-2, doi:10.1007/s12021-018-9360-6 (2018).
2. Gallimore, A. R., Kim, T., Tanaka-Yamamoto, K. & De Schutter, E.: Switching On Depression and Potentiation in the Cerebellum. Cell Reports 22, 722-733, doi:10.1016/j.celrep.2017.12.084 (2018).
3. Myung, J. & Pauls, S. D. Encoding seasonal information in a two-oscillator model of the multi-oscillator circadian clock. European Journal of Neuroscince epub 2017, 48, 2718-2727, doi:10.1111/ejn.13697 (2018).
4. Myung, J., Schmal, C., Hong, S., Tsukizawa, Y., Rose, P., Zhang, Y., Holtzman, M. J., De Schutter, E., Herzel, H., Bordyugov, G. & Takumi, T.: The choroid plexus is an important circadian clock component. Nature Communications 9, 1062, doi:DOI: 10.1038/s41467-018-03507-2 (2018).
5. Nanda, S., Chen, H., Das, R., Bhattacharjee, S., Cuntz, H., Torben-Nielsen, B., Peng, H., Cox, D. N., De Schutter, E. & Ascoli, G. A.: Design and implementation of multi-signal and time-varying neural reconstructions. Scientific Data 5, 170207, doi:10.1038/sdata.2017.207 (2018).
6. Schmal, C., Myung, J., Herzel, H. & Bordyugov, G. Moran's I quantifies spatio-temporal pattern formation in neural imaging data. Bioinformatics 33, 3072-3079, doi:10.1093/bioinformatics/btx351 (2017).
7. Sudhakar, S. K., Hong, S., Raikov, I., Publio, R., Lang, C., Close, T., Guo, D., Negrello, M. & De Schutter, E.: Spatiotemporal network coding of physiological mossy fiber inputs by the cerebellar granular layer. PLoS Computational Biology 13, e1005754, doi:10.1371/journal.pcbi.1005754 (2017).
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
Oral Presentations
1. Hong, S.: Multiplexed coding by cerebellar Purkinje neurons. KAIST, Daejon, South Korea (2017).
2. Hong, S.: Spatiotemporal network coding of physiological mossy fiber inputs by the cerebellar granular layer. KIST, Seoul, South Korea (2017).
3. Hong, S.: Impact of Neuronal Properties on Network Coding. NCTS 2018 Winter school, Frontiers of complex systems science: soft matter, biophysics, and statistical physics, Institute of Physics, Academia Sinica, Taipei, Taiwan (http://phys.cts.nthu.edu.tw/actnews/content.php?Sn=386) (2018).
Conference Invited lectures
1. De Schutter, E.: Molecular models of the early and late phases of bidirectional plasticity at cerebellar synapses. Plenary lecture at CNS*2017, Antwerp, Belgium (2017).
2. Han, D. & Hong, S.: Heterogeneous layers stabilize propagation of a multiplexed spike signal in a feedforward network. CNS*2017, Antwerp, Belgium (2017).
Poster Presentations
1. Bratby, P. & De Schutter, E.: A conductance-based model of cerebellar molecular layer interneurons. CNS*2017, Antwerp, Belgium (2017).
2. Chen, W., Hepburn, I., Casalegno, F., Devresse, A., Ovcharenko, A., Pereira, F., Delalondre, F. & De Schutter, E. STEPS 3: integrating stochastic molecular and electrophysiological neuron models in parallel simulation. CNS*2017, Antwerp, Belgium (2017).
3. De Schutter, E., Hepburn, I., Chen, W., Casalegno, F., Devresse, A., Pereira, F. & Delalondre, F.: Whole cell 3D stochastic molecular simulation with STEPS. Society for Neuroscience, Washington DC, USA (2017).
4. Gallimore, A. R. & De Schutter, E.: An Ultrasensitive ON/OFF Switch Mechanism Controls the Early Phase of Cerebellar Plasticity. CNS*2017, Antwerp, Belgium (2017).
5. Hong, S., Han, D. & De Schutter, E.: Heterogeneous layers stabilize propagation of a spike signal in a feedforward network. Society for Neuroscience, Washington DC, USA (2017).
6. Klingbeil, G. & De Schutter, E.: The use of hardware accelerators in the STochastic Engine for Pathway Simulation (STEPS). CNS*2017, Antwerp, Belgium (2017).
7. Zamora, C. & De Schutter, E.: A model of CaMKII sensitivity to the frequency of Ca2+ oscillations in Cerebellar Long Term Depression. CNS*2017, Antwerp, Belgium (2017).
8. Zang, Y. & De Schutter, E.: Exploring the response to climbing fiber input in Purkinje neurons by a new experimental data based model. CNS*2017, Antwerp, Belgium (2017).
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 OIST Computational Neuroscience Course 2017
- Date: June 26- July 13, 2017
- Venue: OIST Seaside House
- Organizers: Erik De Schutter (OIST), Kenji Doya (OIST), Bernd Kuhn (OIST), and Jeff Wickens (OIST)
- Speakers:
- Upinder Bhalla (NCBS, India)
- Erik De Schutter (OIST)
- Kenji Doya (OIST)
- Bard Ermentrout (University of Pittsburgh, USA)
- Ila Fiete (University of Texas Austin, USA)
- Sonja Grün (Research Centre Jülich, Germany)
- Nicholas Hatsopoulos (University of Chicago, USA)
- Shin Ishii (Kyoto University, Japan)
- Jason Kerr (MPI Bonn, Germany)
- Bernd Kuhn (OIST)
- Sukbin Lim (NYU Shanghai, China)
- Michele Migliore (Institute of Biophysics, Italy)
- David Redish (University of Minnesota, USA)
- Greg Stephens (OIST)
- Yoko Yazaki-Sugiyama (OIST)
- Charles Wilson (University of Texas San Antonio, USA)
7. Other
Nothing to report.



