FY2013 Annual Report
Cell Signal Unit
Professor Tadashi Yamamoto
Abstract
The Cell Signal Unit studies various molecular and cellular events that are important for maintaining healthy life in response to environments. The Unit explores the cause of various diseases that include cancer, neuronal disorder, immunological diseases, diabetes/obesity at the molecular level. More specifically, the Unit characterizes molecular biologically a set of proteins and microRNAs that regulate and execute degradation of mRNAs to silence gene expression. The Unit also studies the mechanisms by which protein kinases and cell adhesion molecules control the brain function such as emotions, learning and memory.
1. Staff
- Dr.Toru Suzuki, Group leader
- Dr.Naosuke Hoshina, Researcher
- Dr.Yo-taro Shirai, Researcher
- Dr.Taku Kureha, Researcher
- Dr.Akinori Takahashi, Researcher
- Ms.Miyuki Hoshina, Technical Staff
- Ms.Chisato Kikuguchi, Technical Staff
- Ms.Xue Li, Technical Staff
- Ms.Miho Tokumasu, Technical Staff
- Ms.Kaori Yamashiro, Research Administrator
2. Collaborations
- Theme: Physiological studies of the CCR4-NOT complex
- Type of collaboration: Joint research
- Researchers:
- Kuba K and Imai Y. Department of Physiology, Graduate School of Medicine, Akita University
- Theme: Structural analysis of the CCR4-NOT complex
- Type of collaboration:Joint research
- Researchers:
- Bartlam M and Rao Z. College of Life Sciences, Nankai University, China
- Theme: Molecular mechanism of mRNA degradation mediated by the CCR4-NOT deadenylase
- Type of collaboration: Joint research
- Researchers:
- Morita M, Fabian M, and Sonenberg N. Department of Biochemistry and Goodman Cancer Research Center, McGill University, Canada
- Theme: Bioinformatics of gene expression affected by impairment of mRNA degradation machinery
- Type of collaboration: Joint research
- Researchers:
- Nagashima T and Okada M. Laboratory for Cellular System Modeling, RIKEN Research Center for Allergy and Immunology
- Theme: Roles of the Fyn tyrosine kinase and its substrates in the central nervous system
- Type of collaboration: Joint research
- Researchers:
- Nakazawa T and Kano M. Department of Neurophysiology, Graduate School of Medicine, University of Tokyo
-
Theme: Roles of the CNOT3 subuni of the CCR4-NOT complex in B cell development
- Type of collaboration: Joint research
- Researchers:
- Inoue T and Kurosaki T. Immunology Frontier Research Center, Osaka University
3. Activities and Findings
Our current interest is to characterize cell signaling that are relevant to physiological properties of mammals such as cancer development, diabetes, immunity and neuronal function.
- The biological role of Tob family proteins and CCR4-NOT complex.
(i) Studies on the Tob family of proteins.
In the study to dissect signaling pathway downstream of ErbB/EGF receptor family tyrosine kinases, we identified the Tob family of proteins, Tob, Tob2, and ANA, that are associated with anti-proliferative function. Upon EGF stimulation MAP kinase became activated downstream of the EGF receptor. We found that MAP kinase in turn phosphorylated Tob, resulting in suppression of the antiproliferative activity of Tob. Further analyses of Tob family proteins with gene manipulated mouse lines revealed followings. #1: Mice lacking Tob are prone to develop cancer. #2: Cells from mice lacking Tob are sensitive to DNA damage-induced apoptosis. #3: Mice lacking Tob show osteopetrosis-like phenotype due to enhanced differentiation and proliferation of osteoblasts. In contrast, #4: mice lacking Tob2 have decreased bone mass due to the increment of osteoclasts. #5: Tob2-deficinet mice exhibit increased adiposity with augmented mass of the epididymal WAT. #6: Mice lacking ana, which is specifically expressed in type II alveolar epithelial cells, develop spontaneous lung adenocarcinoma. The underlying molecular mechanisms of these phenotypes have been addressed to reveal that Tob negatively regulates cyclin D expression. Tob also negatively regulate BMP (bone morphogenetic protein)-mediated signaling. Tob2 negatively regulate formation of osteoclasts by suppressing RANKL expression through its interaction with Vitamin D3 receptor. In addition, Tob2 negatively regulate adipogenesis by inhibiting PPARg2 expression. Finally, clearance of Tob is a novel p53-independent pathway for UV-induced apoptosis. In this mechanism, we found that Cul4-DDB1(Cdt2) induces ubiquitination and degradation of Tob. Interestingly, DNA replication-initiating kinase Cdc7 inhibits Cul4-DDB1(Cdt2)-induced Tob degradation. Cdc7-mediated Tob phosphorylation appears to contribute to suppression of a low dose of UV-induced apoptosis. As Tob is involved in various biological phenomena and thus appears to have different partners to work with in different settings, it is attempting to search for a key partner that plays a relatively common role to control a wide variety of cellular events. Accumulating data from our laboratory and others suggest that CCR4-NOT complex may play such a role.
(ii) Studies on the CCR4-NOT deadenylase complex.
The CCR4-NOT complex consists of at least 10 subunits, CNOT1~3, CNOT6L, CNOT6~11 and is conserved from yeast to human. The complex was initially reported to act as a transcription machinery but later it was found to be associated with the mRNA deadenylase activity. Now we are proposing that the CCR4-NOT complex is important in controlling gene expression circularly, namely at the level of both transcription and mRNA decay (Figure 1).
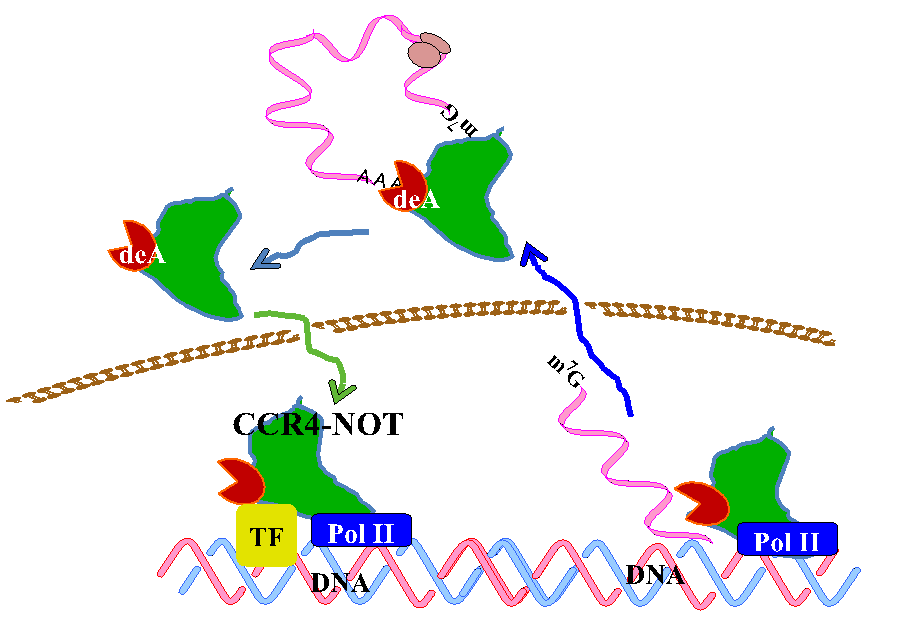
Figure 1 Gene expression is circular
In yeast, two components of the complex, Ccr4p and Caf1p, possesses the deadenylase activity. The mammalian orthologs of Ccr4p are CNOT6 and CNOT6L, and those of Caf1p are CNOT7 and CNOT8. Our recent structural analysis of CNOT6L complexed with nucleotides revealed a deadenylation mechanism involving a penta-covalent phosphate transition. As Tob also interacts with poly(A)-binding protein (PABP), we assume that Tob could be involved in one of the mechanisms by which the CCR4-NOT complex is recruited to the poly(A) tails at the 3’end of mRNAs (Figure 2).
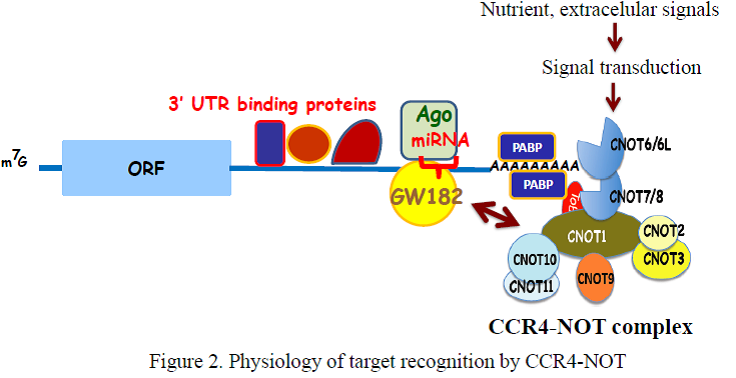
To examine the in vivo function of CCR4-NOT deadenylase complex, we generated gene-engineered mouse lines each lacking individual cnot genes. We found that deficiency of every cnot gene, except cnot6, cnot6L and cnot7, causes embryonic death at distinct developmental stages (Table 1).
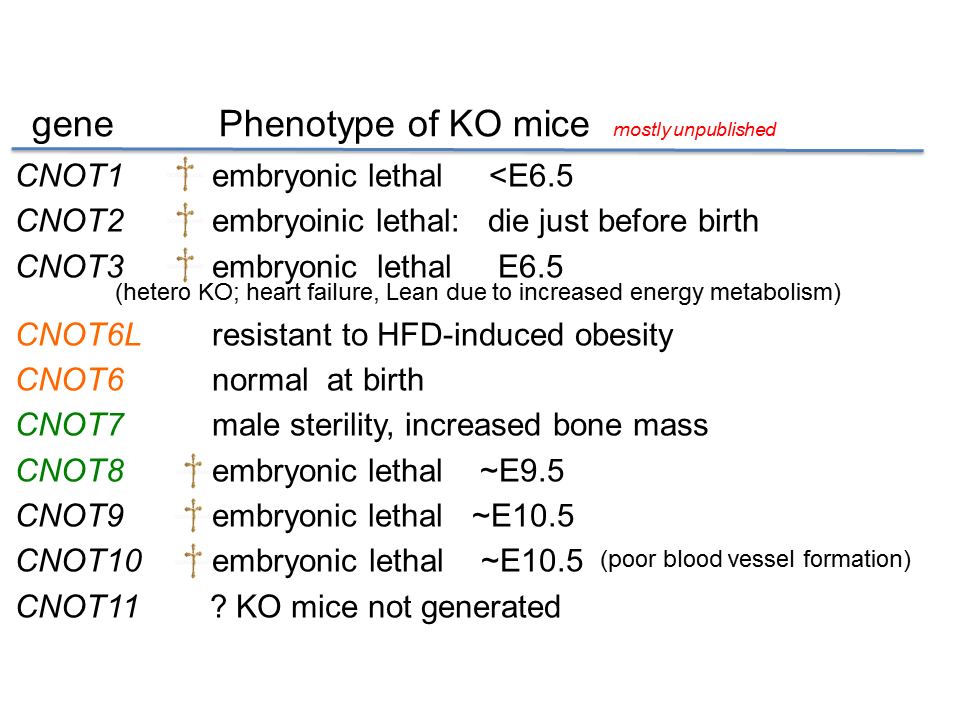
Table 1. Phenotypes of individual Cnot-depleted mice
The cause of death appears to be different between the gene-engineered mouse lines. For example, cnot3-/- mice die due to the defect in early embryogenesis and cnot9-/- mice due to poor vasculogenesis. Though cnot7-/- mice appears normal, male are sterile due to poor spermatogenesis. cnot6L-/- mice are normal except that they are resistant to high-fat diet induced obesity. So far cnot6-/- mice look normal in various analysis except that we haven’t challenged behavioral test.
(iii) Energy metabolism and CCR4-NOT deadenylase complex.
Though cnot3-/- mice are embryonic lethal, cnot3+/- mice are lean with hepatic and adipose tissues containing reduced levels of lipids, and show increased metabolic rates and enhanced glucose tolerance. cnot3+/- mice remain lean and sensitive to insulin even on a high-fat diet. Hepatic expression of most mRNAs is not altered in cnot3+/- vis-à-vis wild-type mice. However, the levels of specific mRNAs, such as those coding for energy metabolism-related PDK4 and IGFBP1, are increased in cnot3+/- hepatocytes. Pdk4 and Igfbp1mRNAs have poly(A) tails that are longer than those seen in control cells. We provided evidence that CNOT3 is involved in recruitment of the CCR4-NOT deadenylase to the 3' end of specific mRNAs.
Similar to cnot3+/- mice, mice deficient in CNOT6L, a catalytic subunit of the CCR4-NOT complex, are resistant to diet-induced obesity and hepatic steatosis and exhibit elevated insulin sensitivity and energy expenditure in a deadenylase activity-dependent manner. Genome-wide search for CNOT6L-associated mRNAs (Figure 3) suggested that the hepatokine Fgf21 mRNA is a possible target of CNOT6L. The hepatokine FGF21 is known to ameliorate diet-induce metabolic disorders such as obesity and hepatic steatosis. Currently we have been trying to clarify a mechanism by which CNOT6L is recruited to the 3’ UTR of Fgf21 mRNA in hepatocytes.
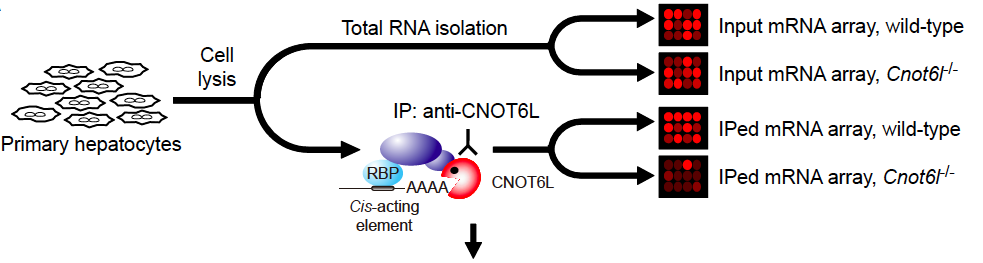
Figure 3 Search for CNOT6L target mRNAs
(iv) Bone and CCR4-NOT deadenylase complex.
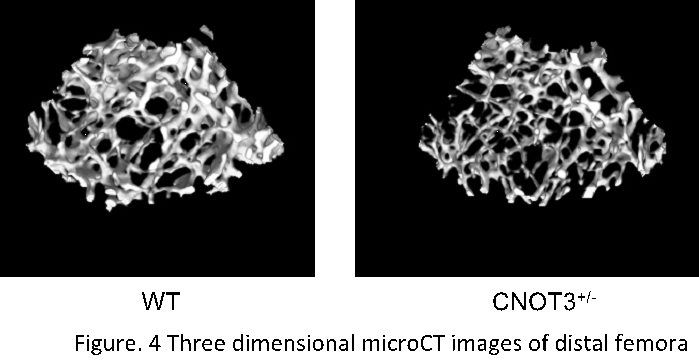
Osteoporosis is a highly prevalent disease affecting nearly 20 million people in the U.S. and is life threatening in elderly patients. Though the steoclastogenesis is under the control of posttranscriptional and transcriptional events, the molecular mechanism is incompletely understood. CNOT3 is a component of CCR4-NOT complex that regulates messenger RNA stability but its function in bone is not known. We have shown that Cnot3 deficiency by a single allele deletion induces osteoporosis. Cnot3 deficiency causes an enhancement in bone resorption in association with an elevation in bone formation resulting in high turn-over type bone loss. At cellular level, Cnot3 deficiency enhances RANKL effects on osteoclastogenesis in a cell autonomous manner. Conversely, Cnot3 deficiency does not affect osteoblasts directly. Cnot3 deficiency does not alter RANKL expression but enhances RANK mRNA expression in bone in vivo. Cnot3 deficiency promotes RANK mRNA stability about two fold in bone marrow cells of the mice. Cnot3 knockdown also increases RANK mRNA expression in the precursor cell line for osteoclasts. Anti-CNOT3 antibody immuno-precipitates RANK mRNA. Cnot3 deficiency stabilizes luciferase reporter expression linked to the 3’-UTR fragment of RANK mRNA. In contrast, Cnot3 overexpression destabilizes the luciferase reporter linked to RANK 3’-UTR. In aged mice that exhibit severe osteoporosis (Figure 4), CNOT3 expression levels in bone are reduced about three fold in vivo. Surprisingly, Cnot3 deficiency in these aged mice further exacerbates osteoporosis and this is also via an enhancement of osteoclastic activity. Our results reveal that CNOT3 is a critical regulator of bone mass acting on bone resorption through at least in part posttranscriptional down regulation of RANK mRNA stability even in aging-induced osteoporosis.
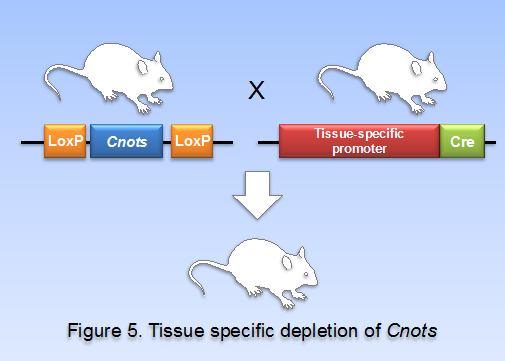
(v) Biological significance of CNOTs in adulthood.
To address the biological significance of CNOT subunits whose un-conditional depletion causes embryonic death, we planned to produce conditional knockout mouse lines by using mice carrying the cre gene under the control of various transcriptional promoters (Figure 5). So far, we have generated mouse lines in which cnot1, cnot3, and cnot8 are individually depleted specifically in the liver (liver-specific KO of cnot1, cnot3, and cnot8, respectively), in the lung (lung-specific KO of cnot3), in the white adipose tissue (WAT-specific KO of cnot3), in the lymphocytes (B cell-specific and T cell specific KOs of cnot3), and in the brain (brain-specific KO of cnot3). All these mouse lines exhibit interesting phenotypes that are under extensive analysis. Preliminarily, we noticed that expression of possible CCR4-NOT target mRNAs is up-regulated in these conditional knock-out mice lines.
It seems that the CCR4-NOT complex targets specific sets of mRNAs for deadenylation depending on a variety of temporal and special conditions. Efforts to elucidate the molecular mechanism by which the CCR4-NOT complex recognizes specific mRNAs are ongoing.
2. Roles of protein-kinase signaling in the central nervous system.
(i) Tyrosine kinase signaling
The Src-family protein-tyrosine kinases are implicated in various neural functions such as formation of neural network, myelination, and synaptic plasticity. We previously showed that GluN2A and GluN2B subunits of NMDA receptors, which play important roles in learning, memory formation, and emotional behavior, are the major substrates of Fyn and Src. We have identified Tyr-1472 phosphorylation on GluN2B and Tyr-1325 phosphorylation on GluN2A as the major tyrosine phosphorylation site of GluN2B and GluN2A, respectively. Further, we have shown that GluN2B Tyr-1472 phosphorylation is important for fear-related learning in the amygdala and that GluN2A Tyr-1325 regulates depression-related behavior.
In parallel of the studies on NMDAR phosphorylation, we have been trying to identify binding partners and substrates of Fyn in the brain. So far we have identified a number of putative mediators of Fyn signaling, including NYAP, FAK, p250GAP, TCGAP, Nogo-A, and RhoGEFs. Recently, we have provided evidence suggesting that TCGAP (=ARHGAP33) is associated with schizophirenia: #1, a single nucleotide polymorphism of TCGAP is associated with decreased expression in patients with schizophirenia; #2, tcgap-/- mice show reduced expression of surface TrkB in neuron, impaired spine development, and deficit in formation of spatial working memory, a phenotype associated with schizophirenia.
We have also searched the data-base for adaptor proteins that may play a part in tyrosine kinase signaling in the central nervous system. We have found an SH2 and SH3 domain-containing Shc-like protein whose biological function haven’t been well characterized. The protein was termed NLNK (neuronal linker protein). We have shown that NLNK expresson is limitted to the central nervous system, especially in the striatum where it interacts with BCR and and with TrkB tyrosine kinase. Physiological and mmolecular biological studies on NLNK are in progress.
(ii) LMTK family signaling
Lemur tyrosine kinases (LMTKs) form a family of protein kinases that are predominantly expressed in the central nervous system (Figure 6).
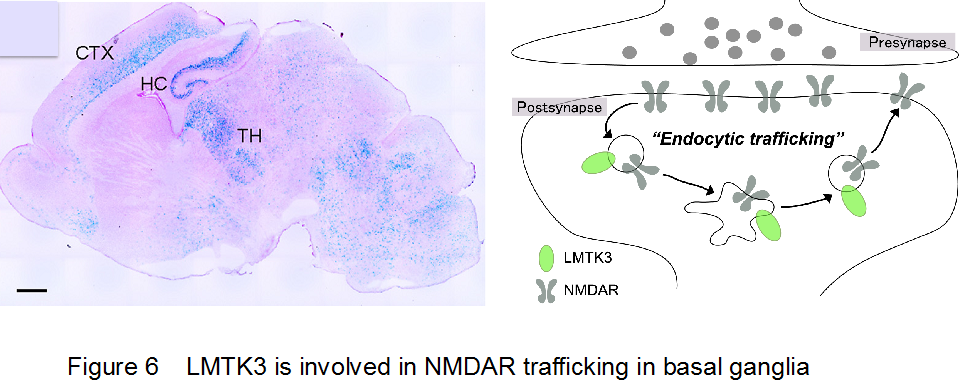
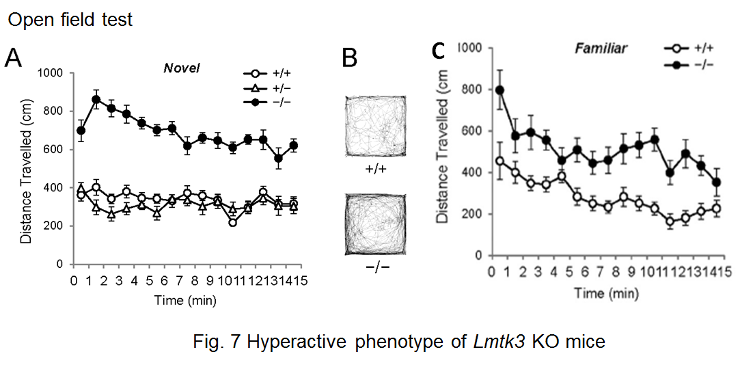
Members of this kinase family, LMTK1-3, comprise an N-terminal transmembrane region, a Ser/Thr kinase domain, and a long C-terminal tail region. We have reported that LMTK2 (or BREK) interacts with a motor protein, myosin VI, and is involved in the regulation of endosomal membrane trafficking in cultured cell lines. LMTK1 also regulates formation of the endocytic regulatory compartment, suggesting that control of membrane traffic is a common function of this family of kinases. Although we have reported that Lmtk2/Brek-deficient mice are infertile and that LMTK2 is essential for the late stages of spermatogenesis, it is not clear how endocytic events are relevant to spermatogenesis. We do not know either the physiological functions of the LMTK2/BREK kinase in the central nervous system. Another member of the LMTK family, LMTK3, is highly expressed in the cerebral cortex, cerebellum, and hippocampus. To address LMTK3’s biological characteristics and in vivo function, we first generated mouse mutants deficient for the lmtk3 gene. We found that lmtk3-/- mice exhibits prominent behavioral abnormalities (Figure 7), including locomotor hyperactivity, reduced anxiety, and decreased depression-like behavior, providing the first evidence for a physiological role of the LMTK family in the central nervous system. Furthermore, we provided evidence that LMTK3 is involved in regulation of endocytic trafficking of NMDAR.
(iii) PCDH17:synaptic connection in basal ganglia
The anatomical topography of neural circuits generally emphasizes distinct functional units. Functional establishment of this topography requires circuit-specific differentiation and refinement of synapses. Synapse development in individual circuits is organized by various types of adhesion molecules. Proteins of the cadherin superfamily, including the protocadherin family participate in synapse-specific interactions. This family of proteins is expressed in synaptic junctions between different types of neurons in neural circuits. Owing to their highly selective adhesive interactions, cadherin-catenin complexes are required for both pre- and postsynaptic development. Although some cadherin members are expressed in specific zones of the basal ganglia, their roles in circuit-specific synaptic development and their physiological significance remain unclear. We have addressed the biological significance of PCDH17, a non-clustered δ2-protocadehrin family member. Our results indicate that PCDH17 plays a crucial role in the regulation of synaptic vesicle assembly in cortico-basal ganglia circuits. Furthermore, PCDH17 deficiency leads to altered presynaptic function in the cortico-striatal pathway. We also observed antidepressant-like phenotypes in PCDH17−/− mice. Further we have shown that expression pattern of PCDH17 is topographycally similar between mouse and monkey. These results provide new insights into the mechanisms underlying the synaptic development of specific cortico-basal ganglia circuits and the physiological role of depression-related behaviors in not only rodents but also in primates.
4. Publications
4.1 Journals
- Hoshina, N., Tanimura, A., Yamasaki, M., Inoue, T., Fukabori, R., Kuroda, T., Yokoyama, K., Tezuka, T., Sagara, H., Hirano, S., Kiyonari, H., Takada, M., Kobayashi, K., Watanabe, M., Kano, M., Nakazawa, T. & Yamamoto, T. Protocadherin 17 regulates presynaptic assembly in topographic corticobasal Ganglia circuits. Neuron 78, 839-854, doi:10.1016/j.neuron.2013.03.031 (2013).
- Watanabe, C., Morita, M., Hayata, T., Nakamoto, T., Kikuguchi, C., Li, X., Kobayashi, Y., Takahashi, N., Notomi, T., Moriyama, K., Yamamoto, T., Ezura, Y. & Noda, M. Stability of mRNA influences osteoporotic bone mass via CNOT3. Proc Natl Acad Sci U S A 111, 2692-2697, doi:10.1073/pnas.1316932111 (2014).
- Shirai, Y.-t. in Recent Progress in Gastric Cancer Vol. 72 最新胃癌学-基礎と臨床の最新研究動向- 637-641 (Nippon Rinsho, 2013).
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- Suzuki, T. Structural basis of the Tob-regulated function of the CCR4-NOT deadenylase complex., in 4th MEXT/Innovative Conference, Seisenryo (2013).
- Takahashi, A. & Morita, M. Functional analysis of the CCR4-NOT complex in obesity, in RNA 2013, Ehime Himegin Hall (2013).
- Takahashi, A. Regulation of obesity by the CCR4-NOT complex, in 1st CCR-4NOT Research Conference, Hotel Grand Tenku (2013).
- Hoshina, N. IFunctional analysis of CCR4-NOT complex in cortical neurogenesis, in 1st CCR-4NOT Research Conference, Hotel Grand Tenku (2013).
- Kureha, T. Functional analysis of CNOT3 which is involved in the differentiation of T cell., in 1st CCR-4NOT Research Conference, Hotel Grand Tenku (2013).
- Shirai, Y. Analysis of Cnot10 KO mice and functional analysis of CNOT3 in lung cancer, in 1st CCR-4NOT Research Conference, Hotel Grand Tenku (2013).
- Suzuki, T. Importance of CCR4-NOT complex-mediated mRNA degradation in liver development, in 1st CCR-4NOT Research Conference, Hotel Grand Tenku (2013).
- Takahashi, A. & Yamamoto, T. Regulation of obesity by the target mRNA of the CCR4-NOT complex, in Signal Network Workshop, Frate Hall, Hokkaido Univeristy (2013).
- Yamamoto, T. Regulation of cell growth and death by Tob protein. Kyoto University Graduate School of Medicine Medical Innovation Center (2013).
- Takahashi, A. Elucidation of obesity regulation by the CCR4-NOT complex, in The 34th Annual Meeting of Japan Society for the Study of Obesity, Tokyo International Forum (2013).
- Yamamoto, T. The dawn of ErbB family oncogene research., in The 36th Annual Meeting of The Molecular Biology Society of Japan, Kobe Port Island (2013).
- Suzuki, T. Importance of CCR4-NOT complex-mediated mRNA degradation in liver development, in The 36th Annual Meeting of The Molecular Biology Society of Japan, Kobe Port Island (2013).
- Hoshina, N. Analysis of the novel adaptor protein in striatal neurons, in The 36th Annual Meeting of The Molecular Biology Society of Japan, Kobe Port Island (2013).
- Kureha, T., Akiyama, T., Morita, M. & Yamamoto, T. Alteration of T cell development and functions in mice with T cell-specific CNOT3 deletion, in The 36th Annual Meeting of The Molecular Biology Society of Japan, Kobe Port Island (2013).
- Shirai, Y. & Yamamoto, T. Functional analysis of CNOT3 in lung cancer, in The 36th Annual Meeting of The Molecular Biology Society of Japan, Kobe Port Island (2013).
- Takahashi, A., Morita, M., Suzuki, T. & Yamamoto, T. Importance of CCR4-NOT deadenylase subunits in obesity, in The 36th Annual Meeting of The Molecular Biology Society of Japan, Kobe Port Island (2013).
- Yamamoto, T. Roles of CNOT3, a component of the CCR4-NOT Deadenylase Complex, in post-transcriptional regulation, NCI, National Institute of Health, USA (2014).
- Yamamoto, T. From tyrosine kinase family of oncogenes to cell signaling., Kobe University (2014).
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Event
6.1 Seminar
- Title: Transgenerational Inheritance of Stress-induced Epigenome Change
- Date: April 1, 2013
- Venue: C016
- Speaker: Shunsuke Ishii
- Chief, Laboratory of Molecular Genetics, RIKEN, Tsukuba
6.2 Seminar
- Title: Novel mRNA and protein quality control systems induced by aberrant translation
- Date: April 8, 2013
- Venue: C015
- Speaker: Toshifumi Inada
- Professor, Graduate School of Pharmaceutical Sciences, Tohoku University
6.3 Seminar
- Title: Quantitative changes in signaling pathway drive qualitative cell decision
- Date: April 24, 2013
- Venue: C016
- Speaker: Mariko Okada-Hatakeyama
-
Team Leader, Laboratory for Integrated Cellular Systems RIKEN, IMS-RCAI
6.4 Seminar
- Title: Cellular and molecular mechanisms for rapid responsiveness of humoral memory responses
- Date: May 24, 2013
- Venue: C016
- Speaker: Tomohiro Kurosaki
- Professor, Laboratory of Lymphocyte Differentiation, WPI Immunology Frontier Research Center, Osaka University
6.5 Seminar
- Title: The role of the WW domain in Golabi syndrome and Hippo tumor suppressor pathway
- Date: June 27, 2013
- Venue: C016
- Speaker: Marius Sudol
- Adjunct Associate Professor, Department of Medicine, Mount Sinai School of Medicine, NYC
6.6 Seminar
- Title: A proteomic approach for identification of the critical proteins that regulate the fate of a specific mRNA and its application to RNA research
- Date: July 8, 2013
- Venue: C016
- Speaker: Shungo Adachi
-
Researcher, Quantitative Proteomics Team, Molecular Profiling Research Center for Drug Discovery (molprof), National Institute of Advanced Industrial Science and Technology (AIST)
6.7 Seminar
- Title: Formation of corticospinal circuitry
- Date: September 9, 2013
- Venue: C015
- Speaker: Yutaka Yoshida
-
Researcher, Division of Developmental Biology, Cincinnati Children’s Hospital Medical Center
6.8 Seminar
- Title: Transcriptional regulation of immediate-early gene response by THOC5, a member of TREX (Transcription/export) complex contributes to growth factors/cytokines induced differentiation
- Date: October 7, 2013
- Venue: C016
- Speaker: Teruko Tamura
-
Professor, Hannover Medical School, Germany
6.9 Seminar
- Title: Insights into recruitment of the CCR4-NOT complex by mammalian gene silencing platforms
- Date: December 2, 2013
- Venue: C016
- Speaker: Marc Fabian
-
Professor, Departments of Oncology and Experimental Medicine Mcgill University
6.10 Seminar
- Title: Engineering Fat Cell Fate to Fight Obesity: Molecular Control of Brown Adipocyte Development
- Date: January 10, 2014
- Venue: D015
- Speaker: Shingo Kajimura
- Assistant Professor, University of California, San Francisco, UCSF Diabetes Center and Department of Cell and Tissue Biology
6.11 Seminar
- Title: Cancer Genome
- Date: January 14, 2014
- Venue: C016
- Speaker: Jun Yokota
- Senior Group Leader, Genomics and Epigenomics of Cancer Prediction Program
Institute of Predictive and Personalized Medicine of Cancer (IMPPC) Barcelona, Spain
6.12 Seminar
- Title: TGF-β signaling in cancer
- Date: February 26, 2014
- Venue: C016
- Speaker: Kohei Miyazono
- Professor, Department of Molecular Pathology, Graduate School of Medicine, The University of Tokyo
6.13 CCR4-NOT Meeting
- Date: August 9-10, 2013
- Venue: Hotel Grand Tencoo
- Organizer: Yumiko Imai (Akita University), Keiji Kuba (Akita University), Tadashi Yamamoto (OIST)
- The number of Participants: 22
6.14 Workshop Gradients and Signalling: from chemotaxis to development 2013
- Date: November 11-15, 2013
- Venue: Seaside House
- Organizer: Mary Ann Price (OIST), Tadashi Yamamoto (OIST), Caroline Hill (CRUK)
- The number of Participants: 50
6.15 Meeting of JST PRESTO(Sakigake) Desing and Control of Cellular Functions
- Date: November 18-19, 2013
- Venue: C209
- Organizer: JST PRESTO(Sakigake)
- The number of Participants: 52




