FY2011 Annual Report
DNA Sequencing Section
Abstract
The DNA Sequencing Section (hereinafter referred to as SQC) currently located in the Okinawa Health Biotechnology Research and Development Center in Uruma city was established in 2008 to support genome research, providing next-generation sequencing (hereinafter abbreviated as NGS) service to all research units in OIST.
SQC has been equipped with five next-generation sequencing machines so far as below.
|
Type |
Number |
|
Genome Sequencer FLX+ (Roche)* |
3 |
|
Genome Analyzer IIx (Illumina) |
1 |
|
Hiseq2000 (Illumina)* |
1 |
*In FY2011, three Genome Sequencer FLX Titanium have been upgraded to FLX+ (plus) and one of the two Genome Analyzer IIx has been upgraded to Hiseq2000.
1. Staff
Section Leader : Takanari Ichikawa
Staff : Azadeh Seidi
Hiroki Goto
Manabu Fujie
Takeshi Usami (till March 2012)
Yuuya Yokoyama (till July 2011)
Nana Arakaki (from March 2012)
2. Collaborations
Through internal researchers (see 4. Publications)
3. Activities and Findings
3.1 Sequencing services
SQC offered the following various sequencing services.
De novo sequencing
●Re-sequencing
●Transcriptome
●ChIP-seq
●miRNA-seq
●Library preparation
In FY2011, SQC provided different types of library preparation (shotgun, mate-pair (paired-end), mRNA, small RNA, and with/without multiplexing) and sequencing services to Satoh Unit, Goryanin Unit, Yanagida Unit, Mikheyev Unit, Samatey Unit, Brenner Unit, and VanVactor Unit.
3.2 Original activities
SQC actively engaged in improvement of sequencing applications and library preparation protocols to ensure obtaining higher quality sequencing data. Our original approaches described below resulted in a drastic improvement in quality and quantity of the sequencing data.
●Technology fusion
The Roche GS FLX+ system is known to provide long-read, and hence, well-suited for de novo sequencing (Fig.1). The system is designed to sequence DNA, which is captured on beads, and amplified by emulsion PCR. SQC has undertaken de novo sequencings of a wide range of species using this sequencing platform. However, original protocols provided by Roche often resulted in obtaining lower read lengths of DNAs than it is said in their catalog and especially so in particular species (Fig. 2). The performance of the system is highly dependent on the signal intensities from the DNA beads which give rise to an appropriate level of amplification. Over-amplification of DNA result in high signal intensities that lead to signal interference, while insufficient amplification of DNA leads to low signal intensities, both yielding a low sequencing performance. Therefore, it has been speculated that the problem was caused by variations in DNA amounts on beads due to an easy-to-amplify feature of the particular genomic sequence specific to a certain species, and thus produce too high signal intensities. To solve the problem, SQC tried to select beads with appropriate signal intensities prior to the sequencing using cell sorter (MoFlo, Beckman Coulter) which is generally used for cell sorting or genome size measurement. The approach resulted in up to two-fold increase in sequencing data amount (Fig. 3). By the combination of the two technologies of the next generation sequencer and cell sorter, challenging sequencing of particular species became possible, which otherwise would have been problematic.
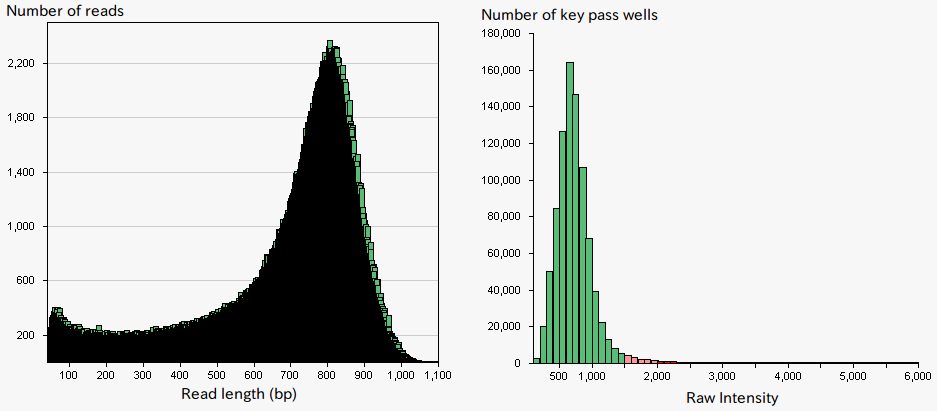
Figure 1:A typical sequencing result and signal intensity
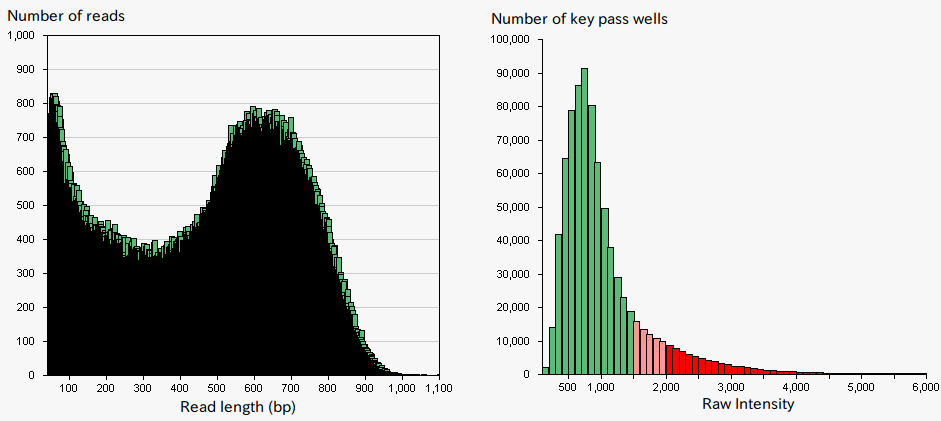
Figure 2:Sequencing result and signal intensity (DNA beads enrichment by Roche protocol)
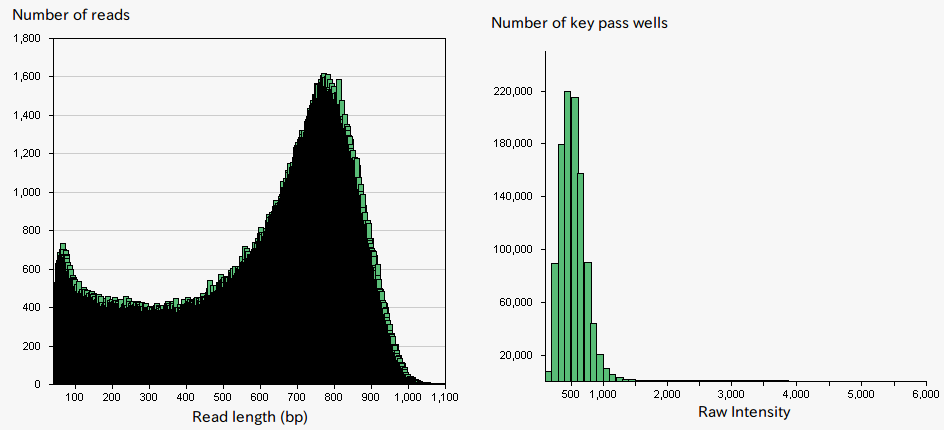
Figure 3:Sequencing result and signal intensity (DNA beads enrichment by cell sorter)
●High throughput library preparation
The Illumina GAIIx and HiSeq2000 systems have a feature to generate the significantly higher number of reads (60 Gb with 300 million reads and 600 Gb with 3 billion reads, respectively, therefore, the sequencing output as well) and are suited for various applications including DNA, RNA, small RNA and ChIP sequencing (e.g. re-sequencing for mutation detection, transcriptome analysis). Taking advantage of these huge capabilities, SQC newly installed the fully-automatic and high-throughput library preparation system consisting of Caliper Life Sciences Sciclone NGS workstation and LabChip GX. The Sciclone NGS workstation is an ultimately flexible but automated liquid handling workstation featuring easy-to-programme new protocol, high accuracy and precision in the motion and is optimized for various NGS applications. The LabChip GX is an automatic gel electrophoresis device that can perform tasks such as sample injection, electrophoresis and fluorescence detection of DNA/RNA band. Moreover, by the automated library preparation system, size and concentration measurement of up to 384 libraries can be handled simultaneously. In SQC, the system is currently applied only for DNA library preparation using TruSEQ DNA Sample Prep Kit, but the system will be used for other available applications in the future.
●Standardization and improvement of library protocols for Next Generation Sequencing
The original reagents and protocols for next generation sequencing differ depending on the manufacturers (Illumina and Roche) even for the same applications. Therefore, it had been difficult to concurrently conduct library preparations for the different sequencers. To shorten the preparation time and to promote standardization of protocols, SQC first tried to standardize a shotgun sequencing protocol which is the most commonly used protocol for the sequencing. It also enabled us to prepare shotgun libraries for different sequencers, and led to reduce the time and cost of library preparation. Moreover, SQC developed a PCR-free shotgun library protocol because PCR amplification lowers the quality of sequencing fidelity. In addition, SQC started to standardize and improve MatePair library preparation critical for de novo sequencing. Those attempts contributed to improvement of N50 contig and scaffold sizes in genome assembly. SQC further extended the attempt to multiplex analysis which is useful for the simultaneous processing of a large number of microorganism genome samples. The application contributed drastically to the improvement of overall output in both Shotgun and MatePair analyses.
3.3 Other activities
●Application Note
SQC provided Nippon Genetics Company with the result using Pippin Prep (automatic DNA size selection system) which was used by the company to prepare a commercial brochure for marketing purposes.
●Technical Exchange Meeting of Okinawa NGS
To collect and exchange information on next generation sequencing techniques and to build a network amongst NGS users in Okinawa, Technical Exchange Meeting of Okinawa NGS was launched in 2011 by Okinawa prefecture (Okinawa Industrial Technology Center and Okinawa Prefectural Agricultural Research Center), the University of the Ryukyus, and OIST The meeting was held four times in FY2011.
4. Publications
4.1 Journals
Takeuchi, T., Kawashima, T., Koyanagi, R., Gyoja, F., Tanaka, M., Ikuta, T., Shoguchi, E., Fujiwara, M., Shinzato, C., Hisata, K., Fujie, M., Usami, T., Nagai, K., Maeyama, K., Okamoto, K., Aoki, H., Ishikawa, T., Masaoka, T., Fujiwara, A., Endo, K., Endo, H., Nagasawa, H., Kinoshita, S., Asakawa, S., Watabe, S., and Satoh N. Draft genome of the pearl oyster Pinctada fucata: A platform for understanding bivalve biology. DNA Research, 19(2): 117-130 (2012).
Shinzato, C., Shoguchi, E., Kawashima, T., Hamada, M., Hisata, K., Tanaka, M., Fujie, M., Fujiwara, M., Koyanagi, R., Ikuta, T., Fujiyama, A., Miller, D. J., and Satoh, N. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature, 476(7360): 320-323 (2011).
Shoguchi, E., Fujie, M., and Hamada, M. No chromosomal clustering of housekeeping genes in the marine chordate Ciona intestinalis. Marine Genomics, 4 (3): 151-157 (2011).
Shoguchi, E., Hamada, M., Fujie, M., and Satoh, N. Direct examination of chromosomal clustering of organ-specific genes in the chordate Ciona intestinalis. Genesis, 49(8): 662-72 (2011).
4.2 Books and other one-time publications
Application note
Yamasaki, S., Usami, T., Goto, H., Seidi, A., and Fujie, M.
Pippin Prep: high reproducibility of DNA sizing
Next Generation Sequencing Catalogue (P13) by NIPPON Genetics Co, Ltd.
4.3 Oral and Poster Presentations
Fujie, M. 454 PTP Re-use. 4th Technical exchange meeting of Okinawa NGS, Okinawa Prefectural Agricultural Research Center, Okinawa, Japan, February 17, 2012
Fujie, M. Construction of biosynthesis gene cluster libraries - Genomic assembly using next generation sequencing technologies -Useful natural compound Production technology development project. 2nd Research and Development Committee, TIME24 Building, Tokyo, Japan. February 7, 2012
Fujie, M. Genome sequencing in OIST "Prospects for Life Innovation by next-generation genome science". Genome Research Seminar sponsored by 164th Committee on Genome Technology, Okinawa Institute of Science and Technology Graduate University, Okinawa, Japan. January 26, 2012
Yamasaki, S. Introduction of DNA sequencing section in OIST. Meeting for technical service with a focus on next generation sequencer, RIKEN Yokohama Institute, Kanagawa, Japan. December 22, 2011
Fujie, M., Usami, T., Koyanagi R., and Satoh, N. Genome sequencing of streptomyces in OIST. Symposium for the project of research hub formulation toward intelligent cluster formation in Okinawa. Okinawa Industry Support Center, Okinawa, Japan, December 20, 2011
Fujie, M. 454 Emulsion Oil. 3rd Technical exchange meeting of Okinawa NGS, Tropical Biosphere Research Center University of the Ryukyus, Okinawa, Japan, November 29, 2011
Usami, T., and Koyanagi, R. Genome sequencing in OIST: Introduction of the next generation sequencing and applications for industry. The Society of Tropical Resources Technologists, Urasoe Industry Promotion Center, Okinawa, Japan. November 25, 2011
Fujie, M. The current status of genome sequencing in OIST -Application of Illumina MatePair for de novo sequencing-. Bioinformatics Training Session (Middle course) organized by Illumina, Inc., Fuji Soft Akiba Plaza, Tokyo, Japan. September 29, 2011
Takeuchi, T., Kawashima, T., Koyanagi, R., Gyoja, F., Fujie, M., Usami, T., Tanaka, M., Ikuta, T., Shoguchi, E., Fujiwara, M., Shinzato, C., Hisata, K., Kinoshita, S., Asakawa, S., Endo, H., Aoki, H., Ishikawa, T., Maeyama, K., Okamoto, K., Masaoka, T., Fujiwara, A., Endo, K., Nagasawa, H., Watabe, S., Nagai, K., and Satoh, N. Draft genome of the Akoya pearl oyster. The Japanese Society of Fisheries Science Autumn Meeting. Nagasaki, Japan, September 29, 2011
Shinzato, C., Shoguchi, E., Kawashima, T., Hamada, M., Hisata, K., Fujie, M., Fujiwara, M., Koyanagi, R., Ikuta, T., Fujiyama, A., Miller, D. J., and Satoh, N. Genome sequencing of the coral Acropora digitifera. Searching for Eve: basal metazoans and the evolution of multicellular complexity, Tutzing, Germany, September 12-15, 2011
Fujie, M. The current status of genome sequencing in OIST –Technological problem of next generation sequencer and its solution- .Okinawa Genome Science Meeting, Okinawa Health Biotechnology Research & Development Center, Okinawa, Japan. September 9, 2011
Yamasaki, S. Digital since early times. Human Resource development Course for Bioinformatics, Okinawa Technology Research Exchange Center, Okinawa, Japan. August 14, 2011
Fujie, M., and Koyanagi, R. Current status of genome research at OIST -Technological improvement and the result-.Genetic Bases for the Evolution of Complex Adaptive Traits, a grant-in-aid for scientific research on innovative areas project, informatics open seminar, Okazaki Conference Center, Aichi, Japan. July 4, 2011
Fujie, M., Shinzato, C., Seidi, A., Usami, T., Yamasaki, S., Satoh, N., Goto, H., and Ichikawa, T. Improved method of Mate-Pair library construction and the its effectiveness in de novo genome assembly. 1st researchers meeting of next generation sequencers. Shizuoka, Japan. May 28-29, 2011
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
1st Technical Exchange Meeting of Okinawa NGS
Yamsaki, S., Goto, H., Seidi, A., Yokoyama, Y., Usami, T., Fujie, M.
Okinawa Industrial Technology Center, Okinawa, Japan, July 19, 2011.
2nd Technical Exchange Meeting of Okinawa NGS
Yamsaki, S., Goto, H., Seidi, A., Usami, T., Fujie, M.
Okinawa Health Biotechnology Research & Development Center, Okinawa, Japan, September 9, 2011.
3rd Technical Exchange Meeting of Okinawa NGS
Usami, T., Fujie, M.
Tropical Biosphere Research Center University of the Ryukyus, Okinawa, Japan, November 29, 2011.
4th Technical Exchange Meeting of Okinawa NGS
Yamsaki, S., Goto, H., Fujie, M., Arakaki, N.
Okinawa Prefectural Agricultural Research Center, Okinawa, Japan, February 17, 2012.
Okinawa Industrial Development Foundation Human Resource Development Project
Human Resource Development Course for Bioinformatics, Next Generation Sequencer Tour
Yamsaki, S., Usami, T., Seidi, A., Goto, H., Fujie, M.
7. Others
Nothing to report.



