【Seminar】Oncolytic Seneca Valley Virus structure and structural insights into receptor specificity
Date
2020年2月19日 (水) 10:30 〜 11:30
Location
C015, Lab1-Level C
Description
Title: Oncolytic Seneca Valley Virus structure and structural insights into
receptor specificity
Speaker: Dr. Nadishka Jayawardena, Department of Microbiology and Immunology, University of Otago
Oncolytic viruses (OVs) are replication competent agents that selectively target cancer cells.
Seneca Valley Virus (SVV) is a newly-discovered oncolytic picornavirus which is classified
as the sole member in the genus Senecavirus. SVV strain 001 has currently completed Phase I
and Phase II clinical trials in pediatric solid tumors and small-cell lung cancer, respectively.
Similar to other picornaviruses, SVV forms naturally occurring empty capsids (without
genome), known as procapsids, which share the same antigenicity as full virions.
Understanding the formation and structure of SVV procapsids could give insights into how to
exploit them as virus-like particles (VLPs) for targeted in vivo drug delivery for cancer
treatment. Recently, we identified the Anthrax Toxin Receptor 1 (ANTXR1), a membrane
protein overexpressed in ~60% of types of cancer, as the high-affinity cellular receptor for
SVV in cancer cells1. However, the high-resolution information on SVV-ANTXR1 interaction
sites remained poorly characterized, thereby hampering the potential to develop SVV mutant
in future oncovirotherapy.
Here, we present how we purified SVV full capsids and procapsids using density gradient
ultracentrifugation and used cryo-electron microscopy (cryo-EM) to solve the structures of full
capsid, procapsid and full capsid-ANTXR1 complex to resolutions of 3.29 Å, 5.9 Å, and 3.8 Å
respectively2,3. Our results show that both full capsids and procapsids have a similar external
structure, while on the interior the main differences were the missing genomic RNA in the
procapsid and a disordered VP1 region. Structural protein analysis in SDS-PAGE revealed the
presence of capsid proteins VP1-VP4 in both full capsids and procapsids. We also show that a
cage of RNA serves to stabilize the inside surface of the full capsid, thereby making it more
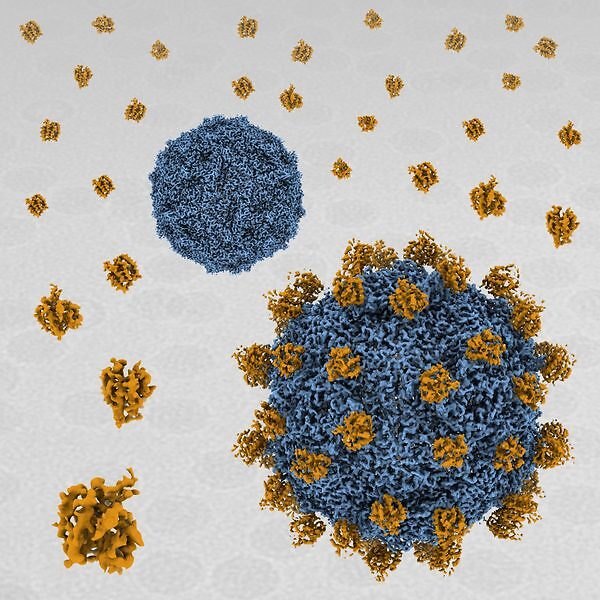
acid stable. In SVV-ANTXR1 complex, ANTXR1 decorates the outer surface of the SVV
capsid and interacts with the surface-exposed BC loop and loop II of VP1, “the puff” of VP2
and “the knob” of VP3. Comparison of the receptor-bound capsid structure with the native
capsid structure reveals that receptor binding induces minor conformational changes in SVV
capsid structure, suggesting the role of ANTXR1 as an attachment receptor. Our results
demonstrate that the capsid footprint on the receptor is not conserved in anthrax toxin receptor
2 (ANTXR2), thereby providing a molecular mechanism for explaining the exquisite
selectivity of SVV for ANTXR1.
Findings from this study lay the foundation for the modification of the SVV procapsid to
develop it for targeted in vivo delivery of therapeutics and to develop potent SVV mutants with
specific cancer tropism.
Seneca Valley Virus (SVV) is a newly-discovered oncolytic picornavirus which is classified
as the sole member in the genus Senecavirus. SVV strain 001 has currently completed Phase I
and Phase II clinical trials in pediatric solid tumors and small-cell lung cancer, respectively.
Similar to other picornaviruses, SVV forms naturally occurring empty capsids (without
genome), known as procapsids, which share the same antigenicity as full virions.
Understanding the formation and structure of SVV procapsids could give insights into how to
exploit them as virus-like particles (VLPs) for targeted in vivo drug delivery for cancer
treatment. Recently, we identified the Anthrax Toxin Receptor 1 (ANTXR1), a membrane
protein overexpressed in ~60% of types of cancer, as the high-affinity cellular receptor for
SVV in cancer cells1. However, the high-resolution information on SVV-ANTXR1 interaction
sites remained poorly characterized, thereby hampering the potential to develop SVV mutant
in future oncovirotherapy.
Here, we present how we purified SVV full capsids and procapsids using density gradient
ultracentrifugation and used cryo-electron microscopy (cryo-EM) to solve the structures of full
capsid, procapsid and full capsid-ANTXR1 complex to resolutions of 3.29 Å, 5.9 Å, and 3.8 Å
respectively2,3. Our results show that both full capsids and procapsids have a similar external
structure, while on the interior the main differences were the missing genomic RNA in the
procapsid and a disordered VP1 region. Structural protein analysis in SDS-PAGE revealed the
presence of capsid proteins VP1-VP4 in both full capsids and procapsids. We also show that a
cage of RNA serves to stabilize the inside surface of the full capsid, thereby making it more
acid stable. In SVV-ANTXR1 complex, ANTXR1 decorates the outer surface of the SVV
capsid and interacts with the surface-exposed BC loop and loop II of VP1, “the puff” of VP2
and “the knob” of VP3. Comparison of the receptor-bound capsid structure with the native
capsid structure reveals that receptor binding induces minor conformational changes in SVV
capsid structure, suggesting the role of ANTXR1 as an attachment receptor. Our results
demonstrate that the capsid footprint on the receptor is not conserved in anthrax toxin receptor
2 (ANTXR2), thereby providing a molecular mechanism for explaining the exquisite
selectivity of SVV for ANTXR1.
Findings from this study lay the foundation for the modification of the SVV procapsid to
develop it for targeted in vivo delivery of therapeutics and to develop potent SVV mutants with
specific cancer tropism.
References
1. Miles LA, Burga LN, Gardner EE, Bostina M, Poirier JT, Rudin CM. Anthrax toxin
receptor 1 is the cellular receptor for Seneca Valley virus. The Journal of Clinical
Investigation. 2017;127(8):2957-2967.
2. Strauss M, Jayawardena N, Sun E, Easingwood RA, Burga LN, Bostina M. Cryo-
Electron Microscopy Structure of Seneca Valley Virus Procapsid. Journal of
Virology. 2018;92(6):e01927-01917.
3. Jayawardena N, Burga LN, Easingwood RA, Takizawa Y, Wolf M, Bostina M.
Structural basis for anthrax toxin receptor 1 recognition by Seneca Valley Virus.
Proceedings of the National Academy of Sciences. 2018;115(46):E10934.
Attachments
All-OIST Category:
Subscribe to the OIST Calendar: Right-click to download, then open in your calendar application.



