FY2017 Annual Report
Evolutionary Neurobiology Unit
Assistant Professor Hiroshi WATANABE

Abstract
The origin of neurons and the evolution of the central nervous system (CNS) are not well understood. In particular, the physiological nature of the first neurons has not been elucidated, and likewise it remains unsolved whether the CNSs of extant bilaterians began with an organized array of nerve net or by a primordial neuronal aggregation. Understanding molecular and cellular features of the nervous systems of the extant basal metazoans - early-branching animal lineages including poriferans, placozoans, ctenophores, and cnidarians - is pivotal to answering these questions. Recent genomic and cellular studies on model basal metazoans have provided new genetic insights into our understanding of the early evolutionary processes of the cellular “neuronalization” and the neural centralization.
Our research projects currently include (1) the anatomical and physiological dissections of the nervous systems of the basal metazoans, and (2) the analysis of genetic mechanism(s) underlying development of the regionalized (semi-centralized) nervous system of cnidarians. Our unit also employs (3) a systematic and comprehensive approaches of neurophysiological features of the basal metazoans. We combine cutting-edge experimental techniques (omics, genetics, and imaging) and informatics technologies (molecular phylogeny and mass spectrometry) to reconstruct the early evolutionary processes of the nervous system.
1. Staff
- Dr. Hiroshi Watanabe, Assistant Professor
- Dr. Eisuke Hayakawa, Group Leader
- Dr. Shinya Komoto, Staff Scientist
- Dr. Mei Fang Lin, Postdoctoral Scholar
- Dr. Amol Dahal, Research Unit Technician
- Ms. Erina Kawai, Research Unit Technician
- Dr. Ryotaro Nakamura, Research Unit Technician
- Ms. Chihiro Kawano, Research Unit Technician
- Mr. Ivan Mbogo, PhD Student
- Ms. Larisa Scheloukhova, PhD Student
- Ms. Christine Guzman, PhD Student
- Mr. Osamu Horiguchi, PhD Student
- Ms. Chihiro Arasaki, Research Unit Administrator
2. Collaborations
Nothing to report.
3. Activities and Findings
3.1 Origin and Early Evolutionary Processes of theNervous System(s).
The origin of the nervous system is one of the most exciting questions in biology. In recent years, thanks to sequencing of genomes of basal metazoans, evolutionary biologists have made spectacular advances in unveiling primitive neuronal components. For example, genes involved in neuronal physiological functions are conserved among bilaterians, cnidarians, and even sponges. The latter possess sensory cells, but not neurons, providing insights into the origin of neurons. Recent findings in the basal metazoans have also raised several important questions, including whether a nervous system arose only once, or multiple times, and whether neural condensations in bilaterian and cnidarian branches reflect a homologous ancestral nature or a paraphyletic neural character. Answers to these questions are pivotal in reconstructing the molecular and cellular features of the nervous systems that existed in ancestral metazoans.
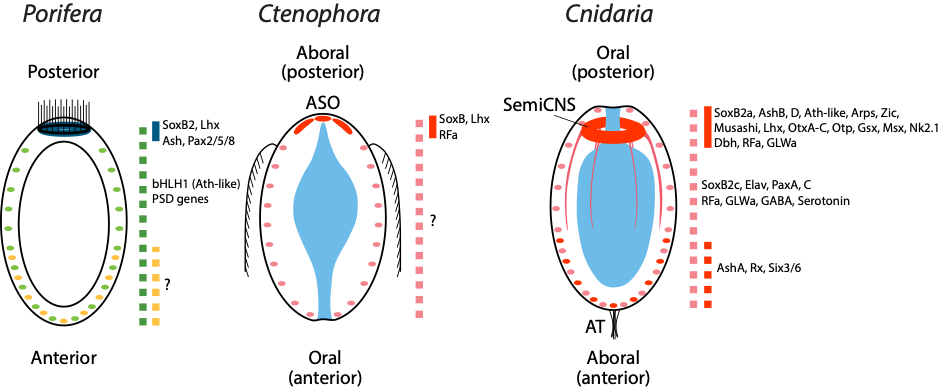
Figure 1: The larval body plans of basal metazoans and expression patterns of neural markers. Regionalized expression of neural marker genes along the primary body axis of three basal metazoan larvae. In Porifera, the blue line indicates neurogenic gene expression in photosensitive pigmented ring cells. The dashed green and dashed yellow lines denote sensory globular cells and flask cells, respectively. The red lines in Ctenophora and Cnidaria indicate posterior neural aggregations from their diffuse nervous systems. The dashed pink line in Cnidaria shows pervasive expression of neural marker genes. Ctenophores possess a diffuse nervous system, whereas no expression of neural marker genes has been shown. The dashed red line in Cnidaria indicates expression of neural markers for aboral nervous system. ASO apical sensory organ, AT apical tuft.
We are now studying 1) genetic repertoires and expression of “neural” genes, 2) neurotransmitters/neuromodulators, and 3) protein machineries such as pre-synaptic and post-synaptic macromolecular complexes to gain functional insights into the ancestral nature of neurons. In order to perform these studies in a systematic way, we are trying with our collaborators to establish constant laboratory cultures of basal metazoan species.
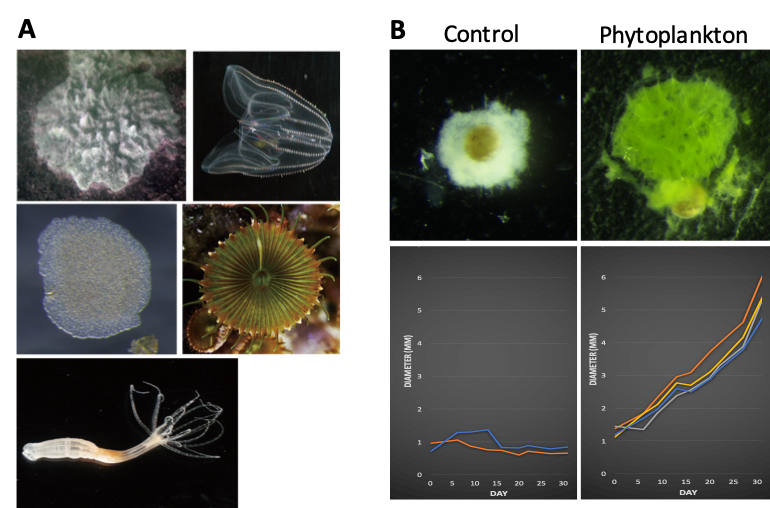
Figure 2: (A) Basal metazoan species in our hand. (B) A stable laboratory culture has so far been established for a poriferan species. This culture system can be run in chemically and biologically clean, allows us to perform the high-quality proteomics/metabolomics analyses in Porifera.
3.2 Systematic Peptidomics of the Basal Animals
In animal kingdom, peptides are widely utilized as intercellular signaling molecules, such as neuropeptides and peptide hormones. They control metabolism, development, behavior and many other biological phenomena as master regulators. Despite their diverse and essential roles in animal biology, it is still largely unknown how the peptide signaling system evolved especially at the early phase of animal evolution.
In FY2017, we continued to work on establishment of analytical system for highly sensitive identification of endogenous peptides. We combined nano-flow liquid chromatography and Orbitrap-based mass spectrometry device. This enabled highly sensitive detection of endogenous peptides, as well as acquisition of their fragment spectra. The acquired fragment spectra were then searched against amino acid sequences derived from genome or transcriptome dataset to identify the structures of endogenous peptides. We applied this analytical workflow to early branching metazoan lineages including Cnidaria, Placozoa, Ctenophora, and Porifera. Computational prediction of peptides, especially short ones, from genomic sequences still remains difficult and not reliable enough for authentic peptide identification. We believe that this approach will enable us to identify novel peptides from the basal animal lineages and open up a new avenue for understanding the complexity of ancestral peptide systems and their physiological functions.
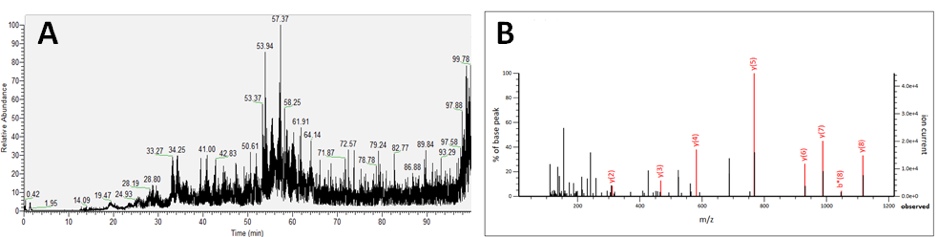
Figure 3: (A) Total ion chromatogram of endogenous peptide extract acquired by liquid chromatography-coupled mass spectrometry. (B) Fragment spectrum and interpretation of fragmentation pattern to deduce peptide structure.
We have so far identified a number of novel peptides from basal metazoans, which have not been predicted from genomic sequences. Immunohistochemical analysis using antibodies that we recently made have revealed many of these peptides are expressed in a specific population of neurons. We are now studying the developmental and anatomical features, and physiological functions of the neuropeptides.
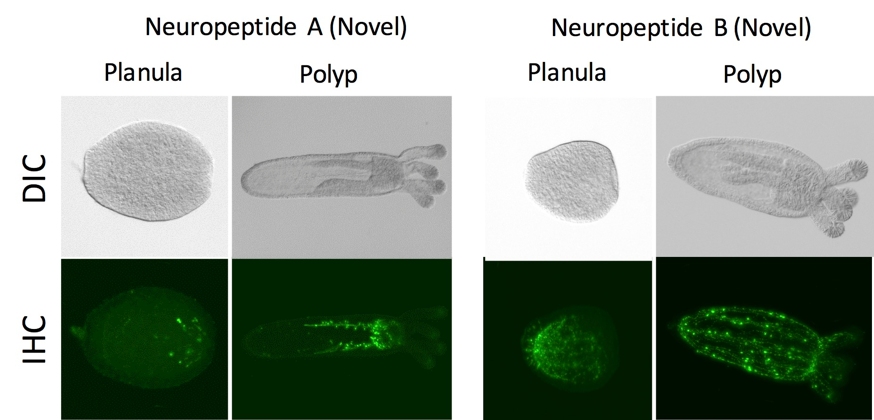
Figure 4: Immunofluorescent images of novel neuropeptides. Our detailed analysis of neuropeptide-expressing neurons in Nematostella (Cnidaria) demonstrated distinct spatial arrangements of neuronal populations and their developmental processes.
4. Publications
4.1 Journals
- Fujimura, Y., Kawano, C., Maeda-Maruyama, A., Nakamura, A., Koike-Miki, A., Yukihira, D., Hayakawa, E., Ishii, T., Tachibana, H., Wariishi, H., and Miura, D. A Chemometrics-driven Strategy for the Bioactivity Evaluation of Complex Multicomponent Systems and the Effective Selection of Bioactivity-predictive Chemical Combinations. Scientific Report. doi:10.1038/s41598-017-02499-1 (2017)
-
Nakamura, J., Morikawa-Ichinose, T., Fujimura, Y., Hayakawa, E., Takahashi, K., Ishii, T., Miura, D., and Wariishi, H. Spatially resolved metabolic distribution for unraveling the physiological change and responses in tomato fruit using matrix-assisted laser desorption/ionization-mass spectrometry imaging (MALDI-MSI). Analytical and Bioanalytical Chemistry. doi: 10.1007/s00216-016-0118-4 (2017)
4.2 Books and other one-time publications
- Watanabe, H. Back Through Time: How Cnidarians and Basal Metazoans Shed Light on Ancient Nervous Systems. Brain Evolution by Design: From Neural Origin to Cognitive Architecture. doi: 10.1007/978-4-431-56469-0_3 (2017)
4.3 Oral and Poster Presentations
- Hayakawa, E. Integration of tandem mass spectrometry, chemo- and network informatics for structural elucidation of small compounds. The 65th Annual Conference on Mass Spectrometry, Japan. Tsukuba, Ibaraki, Japan. May 17 (2017)
- Hayakawa, E., Komoto, S., and Watanabe, H. Novel Neuropeptide Antibodies Visualize The Local Neuronal Networks in Nematostella. International Workshop: The diversification of early emergin metazoans: A Window into Animal Evolution? Tutzing, Germany. September 11-14 (2017)
- Watanabe, H., and Komoto, S. Semi-centralized Nervous System of the Sea Anemone Nematostella Vectensis. International Workshop: The diversification of early emergin metazoans: A Window into Animal Evolution? Tutzing, Germany. September 11-14 (2017)
- Hayakawa, E. Introduction to the informatics aspect of mass spectrometry and metabolomics. The 1st Hackathon for mass spectrometry informatics. Kumamoto, Japan. November 26 (2017)
5. Intellectual Property Rights and Other Specific Achievements
External Fundings
- Grant-in-Aid for Young Scientists B, Japan Society for the Promotion of Science,"Metabolic analysis of neurotransmitters in schizophrenia by ultrahigh-sensitive IM-MS imaging. (超高感度IM-MSイメージングによる統合失調症の神経伝達物質代謝バランス解析)", Lead PI: Hayakawa, E., Amount: 4.29M Yen, Period: Apr. 2017 – Mar. 2019
6. Meetings and Events
6.1 Seminar, Comprehensive understanding of how chromosomes regulate mitosis
Date: September 25, 2017
Venue: OIST Campus
Speaker: Dr. Hideki Yokoyama, Friedrich Miescher Laboratory of the Max Planck Society
7. Other
Nothing to report.



