FY2023 Annual Report
Biological Complexity Unit
Professor Simone Pigolotti

(From left to right) Sharvani Nandikol, Alberto Sassi, Kaori Yamashiro, Simone Pigolotti, Florian Pflug, Robert Ross, Samuel Cure
Abstract
This fiscal year is our first after the unit review, and therefore marks the beginning of a next cycle in our research activity. The most prominent news has been the startup of a small-scale wet lab, which is now complementing our ongoing theoretical research. Such startup has been quite smooth; in particular, the first preliminary experiments have been successful. Aside from this, we are progressing on our research
lines, as outlined below.

Wet lab space of the BCU, 2024
1. Staff
- Prof. Simone Pigolotti, Professor
- Dr. Robert Ross, OIST Interdisciplinary Postdoctoral Fellow
- Dr. Shrabani Mondal, Postdoctoral Scholar
- Dr. Florian Pflug, Postdoctoral Scholar
- Dr. Alberto Sassi, Postdoctoral Scholar
- Dr. Sharvani Nandikol, Postdoctoral Scholar
- Ms. Anzhelika Koldaeva, PhD Student
- Mr. Qiao Lu, PhD Student
- Mr. Samuel Cyrus Cure, PhD Student
- Ms. Kaori Yamashiro, Research Unit Administrator
2. Collaborations
2.1 Cellular energetics
- Description: We are continuing an international collaboration to understand the energy consumption and dissipation of cells using non-equilibrium thermodynamics.
- Type of collaboration: Joint research
- Researchers:
- Pablo Sartori (Gulbekian Institute, Lisbon, Portugal)
- Jonathan Rodenfels (MPI-CBG, Dresden, Germany).
2.4 Error correction in PCR
- Description: We are collaborating with researchers at Tohoku university to understand accuracy of Polymerase Chain Reaction by combining experiments and theory based on non-equilibrium statistical physics.
- Type of collaboration: Joint research
- Researchers:
- Shoichi Toyabe (Tohoku University)
- Hiroyuki Aoyanagi (Tohoku University).
2.5 Martingales in physics
- Description: We have been collaborating with international researchers for a review on the application of the concept of Martingales in theoretical physics.
- Type of collaboration: Joint research
- Researchers:
- Edgar Roldan (ICTP Italy)
- Izaak Neri (King's College London)
- Raphael Chetrite (CNRS Nice)
- Shamik Gupta (TATA Institute Mumbai)
- Frank Julicher (MPIPKS Dresden)
- Ken Sekimoto (ESPCI Paris)
3. Activities and Findings
3.1 Genome replication in asynchronously growing microbial populations
In living cells, DNA replication is carried out by large protein complexes called replisomes. The DNA replication program has been qualitatively studied by looking at the abundance of DNA fragments in an exponentially growing population. We extended our quantitative theory for these patterns to arbitrary microbial populations, including eukaryotes. Our approach was able to successfully infer the location of origins of replications in budding yeast from sequencing data.
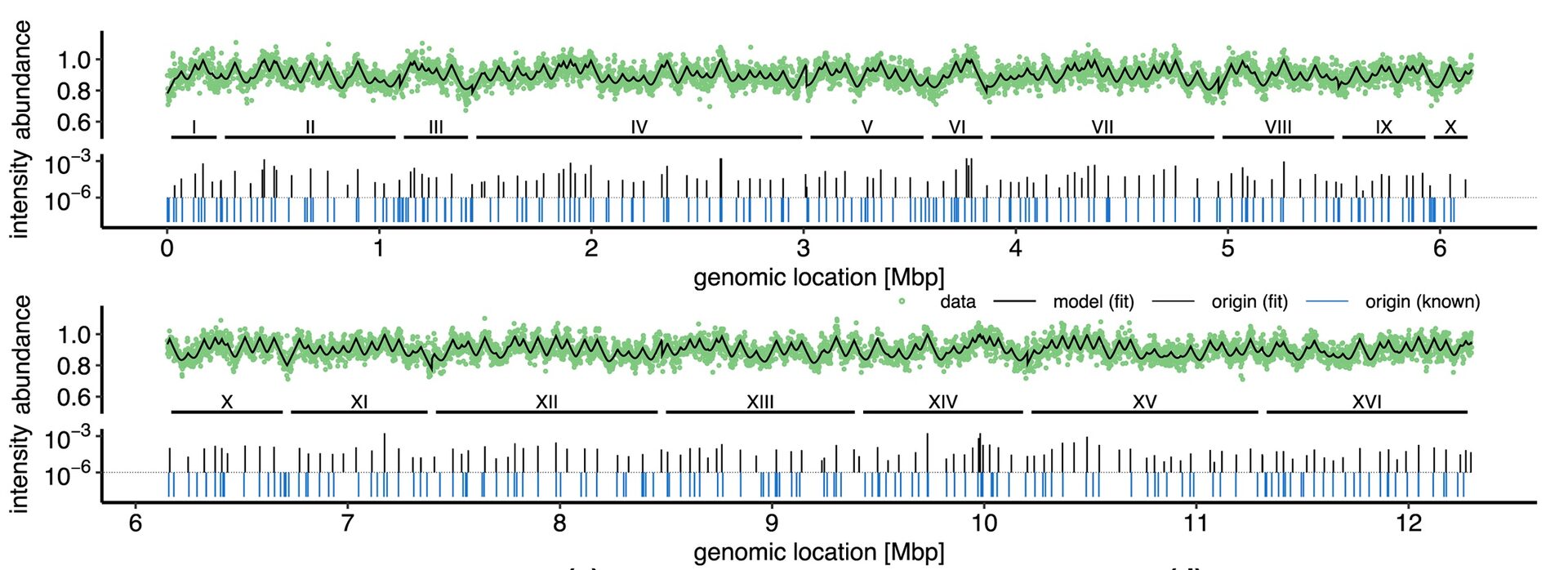
Figure 1: Inferring origins of replication in budding yeast, from Pflug et al. Plos Comp. Biol. 2024.
3.2 Pareto optimal front of kinetic proofreading
Biological information encoded in heteropolymers needs to be replicated at high speed and with a high accuracy, i.e. a low error rate. Thermodynamic analyses of these processes show that the performance of these machines must come at a cost in terms of energy dissipation. However, this tradeoffs are difficult to characterize theoretically. We developed a numerical method based on mathematical optimization theory to compute the optimal solutions in terms of speed, error, and dissipation for the Hopfield model, a paradigmatic example of biological error correction. This project has been finalized and published in New Journal of Physics during this fiscal year.
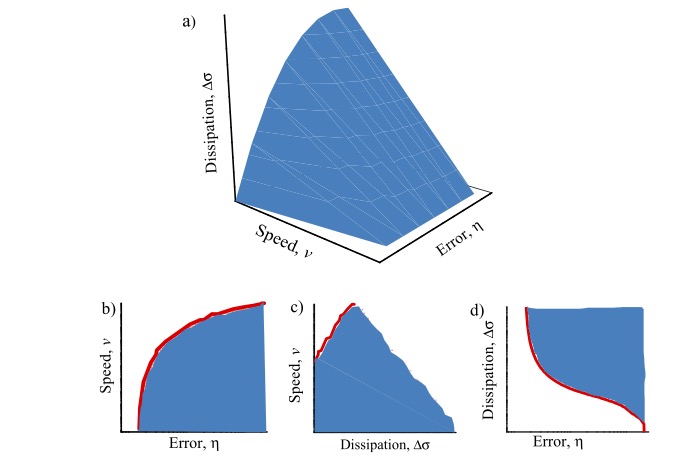
Figure 2: (a) Error-speed-dissipation optimal Pareto front for the kinetic proofreading model. (b) Error-speed Pareto front. (c) Dissipation-speed Pareto front. (d) Error-dissipation Pareto front.
Please use Header 3 or H3 to make subsections of the "Activities and Finding" section, which are H2. How you organize this section is up to you.
3.3 Search dynamics in the CRISPR/Cas9 system
Biological information encoded in heteropolymers needs to be replicated at high speed and with a high accuracy, i.e. This project has been finalized and recently accepted in New Journal of Physics.
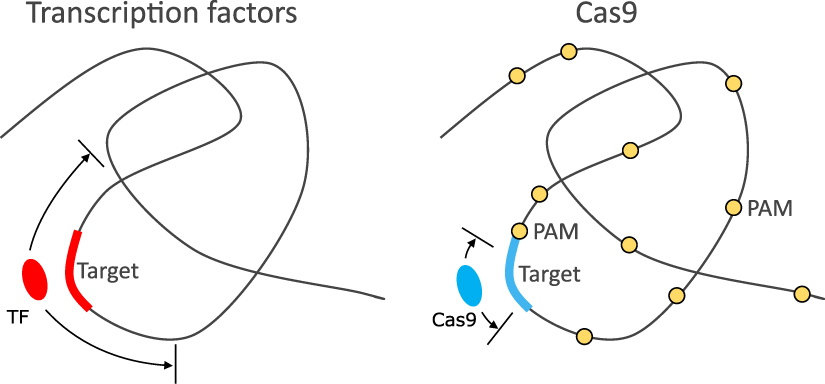
Figure 2: Comparison of target search by a transcription factor and by Cas9, from Lu and Pigolotti NJP 2024.
3.4 Thermodynamic bounds on time-reversal asymmetry
We derived thermodynamic bounds on relative propagators in non-equilibrium systems. These quantities measure the relative probability of reaching a certain state from another one in a given time interval. We show that these relative probabilities are bound by the dissipation, which measure the distance from equilibrium of a system. The results have been published in Physical Review E as a letter.
4. Publications
4.1 Journals
- Q. Lu, S. Pigolotti, "Target search in the CRISPR/Cas9 system: Facilitated diffusion with target cues", accepted in New Journal of Physics (2024).
- É. Roldán, I. Neri, R. Chetrite, S. Gupta, S. Pigolotti, F. Jülicher, K. Sekimoto, "Martingales for physicists: A treatise on stochastic thermodynamics and beyond", arXiv:2210.09983, accepted for publication in Advances in Physics (2024).
- F. Pflug, D. Bhat, S. Pigolotti, "Genome replication in asynchronously growing microbial populations", arXiv:2308:12664, PLOS Computational Biology 20(1), e1011753 (2024).
- S. Liang, S. Pigolotti, "Thermodynamic bounds on time-reversal asymmetry", arXiv:2308.14497, Phys. Rev. E (letter) 108, L062101 (2023).
- D. Chiuchiù, S. Mondal, S. Pigolotti, "Pareto optimal front of kinetic proofreading", arXiv/2208.09381 , New Journal of Physics 25, 043007 (2023).
- G M. Ferlazzo, A M. Gambetta, S. Amato, N. Cannizzaro, S. Angiolillo, M. Arboit, L. Diamante, E. Carbognin, P. Romani, F. La Torre, E. Galimberti, F. Pflug, M. Luoni, S. Giannelli, G. Pepe, L. Capocci, A. Di Pardo, P. Vanzani, L. Zennaro, V. Broccoli, M. Leeb, E. Moro, V. Maglione, G. Martello, "Genome-wide screening in pluripotent cells identifies Mtf1 as a suppressor of mutant huntingtin toxicity", 3962 (2023), Nature Communications (2023)
- E. I. Kiselev, F. Pflug, A von. Haeseler, "Critical Growth of Cerebral Tissue in Organoids: Theory and Experiments", Phys. Rev. Lett. 131, 178402 (2023)
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- S. Pigolotti. Diversity of microbial populations across scales, Centre for Mathematical Biology seminars, University of Bath, UK, May 4, 2023 (2023).
- S. Pigolotti. Speed variations of bacterial replisomes, NCTS STATPHYS28 Satelite Meeting, National Taiwan University, Taiwan, July 31, 2023 (2023).
- S. Pigolotti. Speed variations of bacterial replisomes, OIST - JST Joint Workshop | Biomolecular Super Assembly Insights, Okinawa, Japan, January 19, 2024 (2023)
- S. Pigolotti. Dynamics of genome replication, DPG-Verhandlungen 2024, Berlin, Germany, March 19, 2024 (2023)
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 The SOKOBAN random walk
- Date: August 16, 2023
- Speaker: Dr. Ofek Lauber Bonomo (Tel Aviv University)
7. Other
Nothing to report.



