FY2020 Annual Report
Biological Complexity Unit
Associate Professor Simone Pigolotti
(no photo this year due to social distancing)
Abstract
The Biological Complexity Unit at OIST studies biological and biophysical systems that evolve stochastically. FY2020 has been the fourth year of activity of our Unit, that has been growing in size and productivity. Our main research lines in biophysics are the understanding of non-equilibrium mechanism of error correction in biology and modeling extended microbial populations at individual-based level. A third research line is in statistical physics and in particular in stochastic thermodynamics.
1. Staff
- Prof. Simone Pigolotti, Associate Professor
- Dr. Deepak Bhat, Postdoctoral Scholar
- Dr. Davide Chiuchiu, Postdoctoral Scholar
- Dr. Paula Villa Martin, Postdoctoral Scholar
- Dr. Robert Ross, Postdoctoral Scholar
- Mr. Luke Carter, PhD Student
- Ms. Anzhelika Koldaeva, PhD Student
- Mr. Lu Qiao, PhD Student
- Mr. Shuhei Hara, Rotation Student
- Mr. Tatsuki Hamamoto, Rotation Student
- Ms. Miwa Matsui, Research Unit Administrator (-June 2020)
- Ms. Kaori Yamashiro, Research Unit Administrator
2. Collaborations
2.1 Microbial diversity
- Description: We have been collaborating with the Evolutionary Genomics Unit at OIST to understand the diversity of microbial populations. One first project, lead by our Unit, aimed at explaining the difference in biodiversity between marine and freshwater protists in terms of marine currents This paper was published in Science Advances in this FY. Two other projects, lead by the Evolutionary Genomic Unit, dealt with genomic diversity of gut endosymbionts in termites. One led to a publication in Current Biology in FY2020, whereas the second project was completed in FY2021.
- Type of collaboration: Joint research
- Researchers:
- Prof. Tom Bourguignon, OIST
- Dr. Ales Bucek, OIST
- Dr. Yukihiro Kinjo, OIST
2.2 Stochastic thermodynamics
- Description: We are studying novel universal properties of non-equilibrium thermodynamic systems. During this FY, we characterized thermodynamic uncertainty relations using a variational principle. Together with Luca Peliti, we also wrote a pedagogical book on Stochastic Thermodynamics, that was accepted for publication by Princeton University Press.
- Type of collaboration: Joint research
- Researchers:
- Dr. Izaak Neri, King's College London.
- Dr. Edgar Roldan, ICTP, Trieste.
- Prof. Frank Jülicher, MPIPKS, Dresden.
- Dr. Daniel Busiello, EPFL, Lausannne.
- Prof. Carlos Fiore, University of Sao Paulo, Brazil
- Prof. L. Peliti, SMRI, Italy
2.4 Bacterial growth in microchannels
- Description: We are collaborating with the Micro/Bio/Nanofluidics Unit at OIST to understand bacterial growth in microchannel from a theoretical and experimental perspective.
- Type of collaboration: Joint research
- Researchers:
- Prof. Amy Shen (OIST)
- Dr. Hsie-Fu Tsai (OIST)
3.3 Cellular energetics
- Description: We started an interdisciplinary collaboration at the Kavli Institute for Theoretical Physics to understand the energy consumption and dissipation of cells using non-equilibrium thermodynamics
- Type of collaboration: Joint research
- Researchers:
- Prof. Matthias Heinemann (University of Groningen, Netherlands)
- Pablo Sartori (Gulbekian Institute, Lisbon, Portugal)
- Dr. Jonathan Rodenfels (MPI-CBG, Dresden, Germany).
3. Activities and Findings
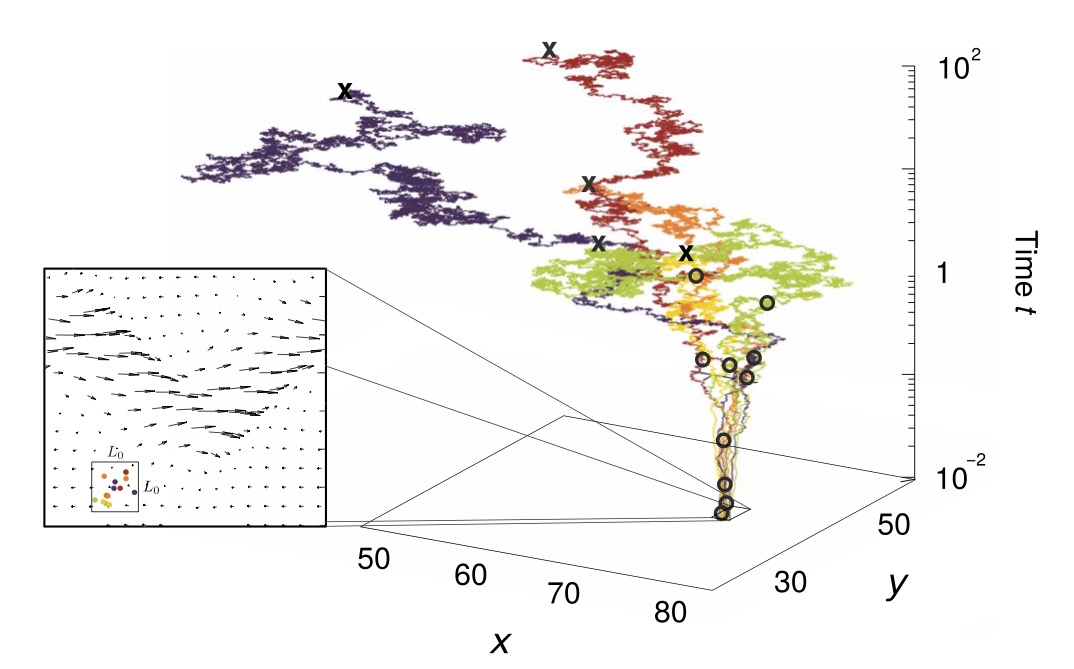
3.1 Ocean currents promote species diversity in protists
We developed an coalescence model to describe the population dynamics of competing microbial species in aquatic environment. Our model predict that, in the presence of transport by oceanic currents, the species-anundance distribution should be characterized by a steepest decay, signaling a more diverse ecosystem. A comparison of metagenomic data from oceans and freshwater is in quantitative agreement with our theoretical prediction (P. Villa-Martin, A. Bucek, T. Bourguignon, S. Pigolotti, Science Advances, 2020).
3.2 Search and localization dynamics of the Crispr/Cas9 system
We propose a stochastic model to understand the sliding dynamics of the CRISPR/Cas9 system. Our model is in quantitative agreement with results from single-molecule experiments. By using an analogy with Anderson localization in condensed matter physics, we characterized localization of Cas9 on a long random stretch of DNA (Lu, Bhat, Stepanenko, Pigolotti, submitted, 2021).
3.3 Population genetics in microchannels
Many microbial population inhabit small channels in porous media. In these channels, reproducing individuals push each other, resulting in expulsion of other individuals from the channel. We developed a theory for the population dynamics of these systems. Our work was performed in collaboration with Shen Unit at OIST, that tested our predictions using microfluidic devices (Koldaeva, Tsai, Shen, Pigolotti, submitted, 2021).
3.4 Generalized Euler-Lotka equation for correlated cell divisions
The growth rate of unicellular populations is related with the distribution of the cell division times by the Euler-Lotka equation, which is a classic result in mathematical biology. However, in the biologically relevant case in which cell division times of mothers and daughters are correlated, the Euler-Lotka equation only provides an approximate result. Using large deviation theory, we derived a generalization of the Euler-Lotka equation which holds for correlated cell divisions (Pigolotti, accepted for publication in Phys. Rev. E (Letter), 2021).
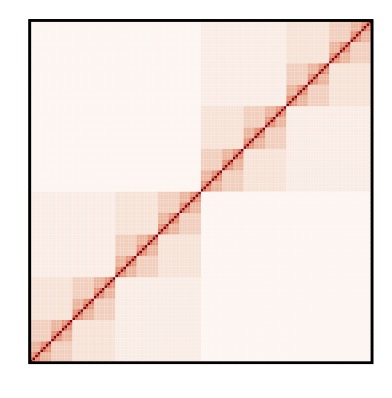
3.5 Bifractal nature of chromosome contact maps
Modern techniques such as Hi-C permit to measure the probability that two chromosomal regions are close in space. Such measurements are advancing our understanding of chromatin structure. We analyzed Hi-C maps using tecniques borrowed from fractal theory. We found that the Hi-C maps of many higher organisms are characterized by two distinct fractal dimensions. We explained this observation using a hierarchical model that we solved exactly. Our results provide a method to analyze Hi-C maps, in particular during organism development (S. Pigolotti, M.H. Jensen, Y. Zhan, G. Tiana, Physical Review Research, 2020).
4. Publications
4.1 Journals
- A. Koldaeva, H. Tsai, A.Q. Shen, S. Pigolotti, "Population genetics in microchannels", arXiv/2104.04161 (2021).
- Q. Lu, D. Bhat, D. Stepanenko, S. Pigolotti, "Search and localization dynamics of the CRISPR/Cas9 system", arXiv/2103.10667 (2021).
- J.M. Miotto, S. Pigolotti, A.V. Chechkin, S. Roldán-Vargas, "Length scales in Brownian yet non-Gaussian dynamics", arXiv/1991.07761 , accepted for publication in Phys. Rev. X (2021).
- S. Pigolotti, "Generalized Euler-Lotka equation for correlated cell divisions", arXiv/2101.02403 , accepted for publication in Phys. Rev. E (Letter), (2021).
- S. Pigolotti, D.Chiuchiù, P.V. Martín, D. Bhat, "Modeling the COVID-19 epidemic in Okinawa", working manuscript (2020).
- S. Pigolotti, M.H. Jensen, Y. Zhan, G. Tiana, "Bifractal nature of chromosome contact maps", Physical Review Research 2, 043078 (2020).
- P.V. Martín, Virginia Domínguez-García, Miguel A Muñoz, "Intermittent percolation and the scale-free distribution of vegetation clusters", New Journal of Physics Vol. 22 (2020).
- P.V Martín, A. Bucek, T. Bourguignon, S. Pigolotti, "Ocean currents promote rare species diversity in protists", Science Advances 6(29), eaaz9037 (2020).
-
T. Bourguignon, Y. Kinjo, P. Villa-Martin, N.V. Coleman, Q. Tang, D.A. Arab, Z. Wang, G. Tokuda, Y. Hongoh, M. Ohkuma, S. Ho, S. Pigolotti, N. Lo, "Increased Mutation Rate is Linked to Genome Reduction in Prokaryotes", Current Biology D-20-00217 (2020).
- J. Bechoefer, S. Ciliberto, S. Pigolotti, É. Roldán, "Stochastic Thermodynamics: Experiments and Theory" (editorial), J. Stat. Mech. 064001 (2020)
4.2 Books and other one-time publications
1. L. Peliti, S. Pigolotti, "Stochastic Thermodynamics: An Introduction". Princeton University Press, in press (2021).
4.3 Oral and Poster Presentations
- Pigolotti, S., "Thermodynamics of Error Correction", ICTP (International Centre for Theoretical Physics) (Trieste, Italy), invited remote seminar, 29 September 2020.
- Pigolotti, S, "Generic Properties of stochastic entropy production", invited remote seminar, Gran Sasso Science Institute (GSSI), L'Aquila, Italy.
- Pigolotti, S., "Thermodynamics of Error Correction", Max-Planck Institute for Dynamics and Self-Organization (Gottingen, Germany), invited remote seminar, 12 January 2021.
- Pigolotti, S., "Coalescent trees in the oceans", contributed talk, APS March Meeting, 17 March 2021.
- Chiuchiu, D., "Error-speed correlations in biopolymer synthesis", APS March Meeting, 19 March 2021.
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 Seminars
- Energetics of biological systems
- Date: March 4, 2021
- Venue: Online
- Speaker: Dr. Jonathan Rodenfels (MPI-CBG, Dresden, Germany)
- What is typical in microbial communities?
- Date: February 25, 2021
- Venue: Online
- Speaker: Dr. Jacopo Grilli (ICTP, Trieste, Italy)
- Spontaneous Escherichia coli persisters with week-long survival dynamics and lasting memory of a short starvation pulse
- Date: February 18, 2021
- Venue: Online
- Speaker: Dr. Namiko Mitarai (Niels Bohr Institute, Copenhagen, Denmark)
7. Other
Nothing to report.