FY2015 Annual Report
Integrated Open Systems Unit
Professor Hiroaki Kitano
Abstract
Integrated Open Systems Unit has made progress on development of tools to support multilevel biophysiological modeling, simulation, and analyzing dynamical systems. Also we have developed a pancreatic beta cell model, and several other models of physiological functions such as cardiac cells, neurons, and ionic channels.
Our Energy projects includes (1) Open Energy System, (2) Microgrid system for Electric Vehicle using Exchangeable batteries and (3) sustainable living in the Hot-Humid Region.
1. Staff
- Dr. Yoshiyuki Asai, Group Leader
- Dr. Kun-Yi Hsin, Staff Scientist
- Takeshi Abe, Technician
- Kyota Kamiyoshi, Technician
- Taichito Sakagami, Technician
- Miwako Nishimura, Technician
- Ken Kuwae, Technician
- Kenichiro Arakaki, Technician
- Midori Tanahara, Research Unit Administrator
2. Collaborations
2.1 Software platform for systems biology
- Type of collaboration: Research Collaboration
- Researchers:
- Dr. Samik Ghosh, Systems Biology Institute
- Ms. Yukiko Matsuoka, Systems Biology Institute
2.2 Open platform for multi-level modeling and simulation
- Type of collaboration: Research Collaboration
- Researchers:
- Professor Nomura Taishin, Osaka University
- Professor Ken-ichi Hagihara, Osaka University
- Dr. Masao Okita, Osaka University
2.3 Modeling and database with dynamic brain platform
- Type of collaboration: Research Collaboration
- Researchers:
- Professor Hiroaki Wagatsuma, Kyusyu Institute of Technology
- Professor Yoko Yamaguchi, RIKEN
2.4 Neurosignal analysis
- Type of collaboration: Research Collaboration
- Researchers:
- Professor Aleesandro E.P. Villa, University of Lausanne
- Professor Alessandra Lintas, University of Fribourg
2.5 Cloud supporting simulation service; Flint K3
- Type of collaboration: Research Collaboration
- Researchers:
- Dr. Nobukazu Yoshioka, National Institute of Informatics
- Dr. Shigetoshi Yokoyama, National Institute of Informatics
- Dr. Masaru Nagaku, National Institute of Informatics
- Professor Ken-ichi Hagihara, Osaka University
- Dr. Masao Okita, Osaka University
2.6 Modeling of function of pancreatic beta cell and analysis for pancreatitis
- Type of collaboration: Research Collaboration
- Researchers:
- MD. Susumu Shinoura, Chubu hospital
2.7 Discovery of anti-influenza agents and drug
- Type of collaboration: Joint research
- Researchers:
- Yukiko Matsuoka. The Systems Biology Institute, Tokyo, Japan.
- Vipul Gupta. The Systems Biology Institute, Tokyo, Japan.
2.8 Open Energy System Project
- Type of collaboration: Joint Research
- Researchers: SONY CSL researchers
2.9 Microgrid system for Electric Vehicle using Exchangeable batteries
- Type of collaboration: Joint Research
- Researchers: PUES researchers
2.10 The sustainable living in the Hot-Humid Region
- Type of collaboration: Joint Research
- Researchers: Misawa Homes Institute of Research and Development researchers
3. Activities and Findings
3.1 Development of software for multilevel modeling: PhysioDesigner
PhysioDesigner is a software that supports to create computable models of physiological systems with spatiotemporal multiple levels. Models built on PhysioDesigner are written in PHML format, which is an XML based specification taking over from ISML. We have rebranded ISML to PHML in December, 2011. In FY2015, we have released three versions of PhysioDesigner, i.e. ver. 1.3 (July, 2015), 1.3.1 (July, 2015), and 1.4 (February, 2016) at physiodesigner.org.
By the developments conducted in FY2015, time series integration into a mathematical model of physiological function became easier and more effective. Besides, a function to model discrete state transitions was implemented. Using this function, users can develop a model of, for example, open/close state model of ionic channels.
Multiplatform support of PhysioDesigner is one of merit for users to use it. To maintain this feature, Java 8 was supported, and Mac OSX 10.11 and Windows 10 were also supported.
Figure 1: (Left) PhysioDesigner dialog to edit state transition model. (Right) Example of simulation result.
3.2 Development of software for simulations of multilevel physiological models: Flint
Flint is a simulator for a multilevel physiological models described in PHML/SBML. In FY2015, we have released Flint 1.3 and 1.3.1 in July 2015. Then we have released ver. 1.4 in February 2016. Improvements of simulation performance, and improvements of user interface were major development. Flint development project became an open source project managed at GitHub to accelerate development speed.
Platform for analysis tools and chart plotters for time series data was initiated to develop concomitantly with Flint and PhysioDesigner.
Figure 2: Application to graph time series data and analyze
3.3 Development of Flint K3 for cloud and HPC computing
Since the size of models is getting larger and larger nowadays, to enable Flint to work on high performance computers is demanded. We have been developing a Flint server, called Flint K3, working on computer clouds, so that users of PhysioDesigner can immediately send simulation jobs to high performance computing environment even if users do not have any accesses to high performance computers.
So far Flint K3 showed a brief figure with multiple panels of graphs of simulated time series that automatically selected. In this FY, graphing function was enhanced. Now users can interactively select variables to draw graphs. Because the data size that must be sent from the simulation server to the client can be huge, the data is gradually sent, and graph resolution is gradually improved as data points that the client received increase.
Figure 3: Flint K3 parameter set window and interactive chart plot window
3.4 Model development of pancreatic beta cell and analysis for pancreatitis
Pancreatic beta cell plays an important role to control blood sugar level by secreting insulin. Malfunction of insulin secretion of beta cells is one major reason of diabetes. Prediction of dynamics of membrane potential and ions concentration for the cases of normal condition, pathological condition and medication can be beneficial for considering therapeutic strategy and for discovering new drugs. We have initiated a collaboration with Okinawa Prefectural Chubu Hospital on prediction of pathological condition of diabetes, and of symptoms of post-ERCP acute pancreatitis. We have already an acceptance from ethical committee to deal with anonymized patients data provided by Chubu hospital.
Figure 4: Scheme of a beta cell model displayed on PhysioDesigner, and simulation results for normal and pathological cases.
3.5 Development of intelligent screening pipeline for network pharmacology-based prediction.
Drugs may interact with multiple molecules in the human body, and such drug action, known as polypharmacology, may be efficacious or deleterious for the treatment of disease. For example, β-lactams exhibit antibacterial action principally by targeting multiple penicillin-binding proteins. Similarly, a multi-target strategy has advanced the treatment of neurodegenerative diseases. On the other hand, poor drug selectivity may increase therapeutic risks and negatively impact drug development because of unintended drug-target interactions. An example of this is the cardiotoxicity of kinase inhibitor Sunitinib. Identification of drug targets is therefore a critical stage of drug development. Drug target identification is therefore a critical task of drug design.
It is only relatively rare that a promising screening pipeline for prediction of drug-target interactions have become available to research groups. Results from some resources mainly focus on only predicting interaction targets, but network pharmacology information is limited. Advances in systems biology are creating opportunities for the application of network pharmacology to identify drug targets over molecular pathway maps. Many of well-curated pathway maps explicitly describes molecular interactions. These open resources allow a deeper understanding of how small molecules acts within molecular networks.
We have developed an intelligent screening pipeline to make network-based screening efficient and practical. It incorporates an elaborately designed scoring function called docK-IN (combining docking with intelligence) for molecular docking to assess protein-ligand binding potential. docK-IN was initially developed in our previous work to address the critical issue of commonly used docking programs that were too inaccurate for reliable prediction. We have also developed a web service called systemsDock (http://systemsdock.unit.oist.jp/) specifically designed for network pharmacology-based prediction and analysis, which permits docking simulation and molecular pathway map for comprehensive characterization of ligand selectivity and interpretation of ligand action on a complex molecular network. For large-scale screening and ease of investigation, systemsDock has a user-friendly GUI interface for molecule preparation, parameter specification and result inspection. Ligand binding potentials against individual proteins can be directly displayed on an uploaded molecular interaction map, allowing users to systemically investigate network-dependent effects of a drug or drug candidate (Figure 5-1). systemsDock is the first web service enabling drug developers to carry out network pharmacology-based prediction and analysis by integrating results from structural biology with systems biology. We have applied the intelligent screening pipeline together with the web service to drug discovery project practically. For instance, we investigated compounds anti-influenza therapy using systemsDock with a deeply curated influenza infection pathway called FluMap (http://www.influenza-x.org/flumap/). For demonstration, a colored FluMap shows network-based screening results (Figure 5-2).

Figure 5-1. Screenshots of the systemsDock web interface. (A) Interactive functions for binding site specifications are accessed by clicking on the displayed protein structure or amino acids listed in the sequence table to define the location of the preferred binding site. Users can adjust x-y-z coordinates to refine the location. (B) Links are provided for the test compound in external databases, as well as to visualize the compound in 3D. (C) Prediction results are furnished in an interactive histogram. Docking scores for each compound are grouped by proteins. By clicking on one of the bars, molecular binding interactions can be graphically shown in 2D/3D for structure-based investigation as shown in (F). (D and E) Visualizing results through a pathway map provided by the user or using a heat map. Colors of proteins are displayed as white-to-red scales or as white and red according to the docking scores. Click on a colored node (i.e. protein) to display binding interactions in 2D/3D as shown in (F). (F) Visualizing protein-ligand binding interactions of the test compound or native ligand in 2D/3D. Protein residues involved in the binding interaction are automatically identified. For reference, those that interacted with a native ligand, if available, are also listed. Clicking on any of the residue entries listed allows users to center and display the specified residue for closer inspection.
Figure 5-2. Prediction of a test compound (PubChem CID: 3795) against 180 FluMap proteins using systemsDock. FluMap describes the intermolecular network of influenza A virus life cycle from (1) entry, (2) endocytosis, (3) fusion, (4) transcription/translation, (5) assembly, (6) packaging to (7) budding. Docking scores are converted into white/red color according to a cutoff value at 5.52 pKd (white: inactive; red: active; gray: not in the test or no results). Results are projected on the pathway map. The JAK1 protein, which plays a major role in the JAK/STAT signaling pathway, is enclosed in a black frame.
3.6 Open Energy System Project
From December 2014, DCOES are operated in 19 faculty houses in OIST campus and are continuously supplying electricity to houses and collect data. DCOES real data in 2015 show average 141.5 kWh energy generated and supplied to residents. Around 10-17 kWh energy was exchanged per day. Average self-sufficient rate (ratio of supplied energy from solar panels) is 0.37 and max is 0.61.
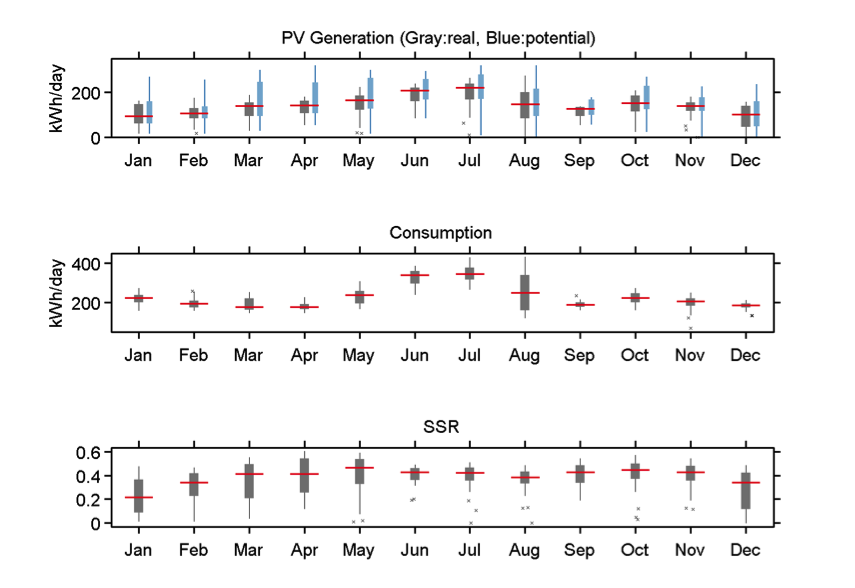
Figure 6: In 2015, DCOES PV generation, Consumption, and SSR
3.7 Microgrid system for Electric Vehicle using Exchangeable batteries
This project is to develop the following three systems; 1) exchangeable batteries, 2) EV for these batteries, and 3) charging station (SwapStation) from renewable energy, such as solar panels. In FY 2015, we finished to develop the first version of those three systems and will confirm how efficiently use the renewable energy with exchangeable batteries comparing existed the EVs and the EV charging stations. Optimal management of exchangeable batteries is one of important knowledge as well from this project. There are two aspects of optimal batteries managements; one is for battery life cycle, and the other is for efficient use of renewable energy.

Figure 7: EV for exchangeable batteries and charging station
3.8 The sustainable living in the Hot-Humid Region
This project aims to provide the highest quality of life in remote areas that lack basic infrastructure (water, energy, etc.). Based on this concept, we finished to build the experimental house with Misawa R&D. This house includes 8 kW PVs and 2 kW wind turbine as electrical generator, 3 kWh battery system, and DC appliance as consumption. In addition to these renewable energy system, the following four systems are built in; 1) Cascade Solar system, 2)Cascade Solar desiccant, 3) Wall radiation cooling system, and 4)Locally supplied water system. We will collect and anlyze data from this experimental house, and would like to suggest the optimal system structure in terms of energy and water.

Figure 8: The experimental house and EV
4. Publications
4.1 Journals
- Amemiya T, Honma M, Kariya Y, Ghosh S, Kitano H, Kurachi Y, Fujita F, Sasaki Y, Homma Y, Abernethy DR, Kume H, Suzuki H. Elucidation of the molecular mechanisms underlying adverse reactions associated with a kinase inhibitor using systems toxicology. npj Systems Biology and Applications. 1, 2015.Asai Y,
- Abe T, Li L, Oka H, Nomura T, Kitano H. Databases for multilevel biophysiology research available at Physiome.jp. Front Physiol. 6: 251, 2015.
- Kawakami E, Singh VK, Matsubara K, Ishii T, Matsuoka Y, Hase T, Kulkarni P, Siddiqui K, Kodilkar J, Danve N, Subramanian I, Katoh M, Shimizu-Yoshida Y, Ghosh S, Jere A, Kitano H. Network analyses based on comprehensive molecular interaction maps reveal robust control structures in yeast stress response pathways. npj Systems Biology and Applications. 2, 2016.
- Kitano H. Accelerating systems biology research and its real world deployment. npj Systems Biology and Applications. 1, 2015.
- Kitano H. Artificial Intelligence to Win the Novel Prize and Beyond: Creating the Engine for Scientific Discovery. AI magazine. 37: 39-49, 2016.
- Lopes TJ, Shoemaker JE, Matsuoka Y, Kawaoka Y, Kitano H. Identifying problematic drugs based on the characteristics of their targets. Front Pharmacol. 6: 186, 2015.
- Nim HT, Furtado MB, Costa MW, Kitano H, Rosenthal NA, Boyd SE. CARFMAP: A Curated Pathway Map of Cardiac Fibroblasts. Plos One. 10, 2015a.
- Nim HT, Furtado MB, Costa MW, Rosenthal NA, Kitano H, Boyd SE. VISIONET: intuitive visualisation of overlapping transcription factor networks, with applications in cardiogenic gene discovery. BMC bioinformatics. 16: 141, 2015b.
- 北野宏明. 人工知能がノーベル賞を獲る日, そして人類の未来−究極のグランドチャレンジがもたらすもの−.人工知能学会誌 Vol. 31 275-286. (2016) [Kitano H. The Day AI Win the Noble Prize and the Future of Humanity –An Ultimate Grand Challenge in AI and Scientific Discovery. Journal of the Japanese Society for Artificial Intelligence Vol. 31 275-286. (2016)]
- 浅井義之, 安倍武志, 北野宏明. PhysioDesignerによる生理機能の多階層モデリング.日本薬理学雑誌 114-119 (2016). [Asai Y, Abe T, Kitano H. Multilevel modeling of physiological functions with PhysioDesigner.Journal of Pharmacological Sciences Vol. 147 114-119 (2016)]
- 浅井義之, 安倍武志, 北野宏明. 生理機能の多階層ハイブリッドモデリングを実現するモデリングプラットフォーム.生物物理 120-124 (2016) [Asai Y, Abe T, Kitano H. Modeling platform for developing multilevel hybrid models of physiological functions. SEIBUTSU BUTSURI Vol. 56. (2016)]
- 浅井義之. 安倍武志, 野村泰伸, 北野宏明. 生体医工学 Vol. 53 144-150 (2015) [Asai Y, Abe T, Nomura T, Kitano H. Software platform for multilevel modeling of physiological systems. Medical and Biological Engineering Vol. 53 144-150 (2015)]
- 北野宏明. システム・トキシコロジーの展開. QIAGEN eyes Vol. 12 7-9 (2015) [Kitano H. Introduction to Systems Toxicology. QIAGEN eyes Vol. 12 7-9 (2015)]
- 北野宏明. システム・トキシコロジーの展開 (第二回). QIAGEN eyes Vol. 13 7-9 (2015) [Kitano H. Introduction to Systems Toxicology (2nd). QIAGEN eyes Vol. 13 7-9 (2015)]
- 坂上太一郎, 浅井義之, 西村美和子, 西銘恵三, 徳田佳一, 川本大輔, 所眞理雄, 北野宏明. 沖縄における直流マイクログリッドシステム実証試験. 太陽エネルギー Vol. 41. 5. 43 (2015) [Sakagami T, Asai Y, Nishimura M, Nishime K, Tokuda Y, Kawamoto D, Tokoro M, Kitano H. in SOLAR ENERGY Vol. 41. 5. 43 (2015)]
4.2 Books and other one-time publications
- Matsuoka Y, Fujita K, Ghosh S, Kitano H. Weaving Knowledge into Biological Pathways in a Collaborative Manner. in Computational Systems Toxicology (eds Hoeng J, Peitsch MC) pp181-208 ISBN: 978-1-4939-2777-7 . (Springer New York, 2015).
4.3 Oral and Poster Presentations
[Oral presentations]
- 北野宏明. システム医科学におけるオープンイノベーションを促進するガルーダ・プラットフォーム. 第23回 分子複合医薬研究会 特別シンポジウム. 産業総合技術研究所 関西センター (大阪府) 2016.02.26. [Kitano H. Garuda Platform for Open Innovations in Systems Medicine. 23rd Molecular Composite Medicine Society Special Symposium. AIST Kansai Center, Osaka JAPAN. 2016.02.26.].
- 北野宏明. システム創薬とデジタルヘルスを促進するガルーダ・プラットフォーム. 第10回スーパーコンピュータ「京」と創薬・医療の産学連携セミナー. グランフロント大阪 (大阪府) 2016.01.22. [Kitano H. Garuda Platform for Systems Drug Discovery and Digital Health. The 10th university-industry research collaboration seminar on Supercomputer K, drug discovery and medicine. Grand Front Osaka, Osaka JAPAN, 2016.01.22]
- 北野宏明. ガルーダ・プラットフォーム:創薬、デジタルヘルスのための革新的情報基盤システム. 武田薬品―京都大学医学部附属病院合同シンポジウム: 日本におけるデジタルヘルス -現状と将来展望. 京都大学医学部創立百周年記念施設芝蘭会館 稲盛ホール (京都) 2015.12.04. [Kitano H. Garuda Platform: Innovations in Compatational Platform for Drug Discovery and Digital Health. Takeda Pharmaceutical Company-Kyoto University Hospital Joint Symposium. Kyoto University, Shiran-Kaikan, Inamori Hall, Kyoto. 2015.12.04.]
- 北野宏明. システム医科学におけるオープンイノベーションを促進するガルーダ・プラットフォーム. 日本薬物動態学会 第30回年会. タワーホール船堀 (東京都) 2015.11.14. [Kitano H. Garuda Platform for Open Innovations in Systems Medicine. 30th JSSX Annual Meeting in Tokyo . Tower Hall Funabori, Tokyo. 2015.11.14.].
- 北野宏明. 人工知能がノーベル賞を獲る日:科学的発見のエンジンを作る. 日本バイオインフォマティクス学会年会・生命医薬情報学連合大会, 一般公開シンポジウム:生命科学データベースと人工知能・ロボティクスの拓く未来. 京都大学宇治キャンパスおうばくプラザ (京都) 2015.10.31. [Kitano H. The Day AI win Nobel Prize. Informatics in Biology, Medicine and Pharmacology 2015, The 2015 Annual Conference of the Japanese Society for Bioinformatics (JSBi 2015). Kyoto University, Uji Campus, Obaku Plaza, Kyoto. 201510.31.]
- 北野宏明. 標準化: VRML, SBML, SBGN, RobCupそしてガルーダプラットフォームを経て. 汎用ヒト型ロボット活用研究会ワークショップ 「汎用ヒト型ロボット活用に向けたデータサイエンス」. クックパッド本社 (東京都) 2015.09.05. [Kitano H. Creating Standards: Experiences from VRML, SBML, SBGN, RoboCup, and Garuda Platform. General-purpose humanoid robot utilization Workshop: Data science for the general-purpose humanoid robot utilization. Cookpad, Inc., Tokyo. 2015.09.05.]
- 北野宏明. システム医科学におけるオープンイノベーションを促進するガルーダ・プラットフォーム. JASIS 2015 先端診断イノベーションゾーン. 幕張メッセ (千葉県) 2015.09.03. [Kitano H. Garuda Platform for Open Innovations in Systems Medicine. JASIS 2015 Advanced Diagnostics, Innovations Zone/JASIS 2015. Makuhari Messe, Chiba, 2015.09.03.]
- 北野宏明. ガルーダプラットフォーム: 創薬およびデジタルヘルスのための計算プラットフォームの革新. 第31回創薬セミナー. 大津プリンスホテル (滋賀県) 2015.07.22. [Kitano H. Garuda Platform: Innovations in Computational Platform for Drug Discovery and Digital Health. The 31st Frontier Seminar for Novel Medicinal Sciences. Otsu Prince Hotel, Shiga. 2015.07.22.]
- 北野宏明. システムトキシコロジーの情報基盤としてのGaruda Platform. 第42回日本毒性学会学術年会 シンポジウム:「エピゲノミクス・ゲノミクス解析の進展と適応拡大する毒性オミクス」. ホテル日航金沢 (石川県) 2015.07.01. [Kitano H. Garuda Platform as a basis for systems toxicology. The 42nd Annual Meeting of the Japanese Society of Toxicology, Symposium: Extending area of Toxicomics in conjunction with progress in epigenomics and genomics analysis. Hotel Nikko Kanazawa, Ishikawa. 2015.07.01.]
- 北野宏明. オープンエネルギーシステム. 気候変動と島しょ:エネルギー自給自足に向けて−日仏討論会. 沖縄科学技術大学院大学 (沖縄). 2015.10.22 [Kitano H. Japanese-French Debate "Islands and Climatic Change: Towards Energy Autonomy" Japanese-French Debate "Islands and Climatic Change: Towards Energy Autonomy". OIST, Okinawa JAPAN. 2015.10.22.]
- Kitano H. Cellular systems. VIZBI 2016. EMBL, Heidelberg, (2016.03.10).
- Kitano H. Computational Systems Toxicology Approach Using the Garuda Platform. Workshop on Pharmacological Mechanism-Based Drug Safety Prediction: Integrated Computational Approaches for Problem Solving of Systems Comprised of Multiple Levels of Biological Organization, from Drug Exposure to Clinical Phenotype . FDA White Oak Facility, Silver Spring, USA (2015.08.06).
- Sakagami T, Asai Y, Werth A, Kawamoto D, Tokoro M, Kitano H. Performance of a DC-Based Microgrid System in Okinawa. International Conference on Renewable Energy Research and Applications(ICRERA). Palermo Italy. 2015.11.23.
- Kitano H. Garuda Platform for Systems Biology. ICSB 2015. Biopolis, Singapore. 2015.11.23.
- Kitano H. Dysregulation of suppressor of cytokine signaling 3 in keratinocytes cause hyper-activation of Ap-1 related genes and develop hyperplasia like phenotype. ICSB 2015. Biopolis, Singapore. 2015.11.23.
- Kitano H. Integrated Open Energy System at OIST. Hawaii – Okinawa Energy Innovation Forum. Chamber of the House of Representatives, Hawaii State Capitol, Hawaii, USA. 2015.07.10.
- Kitano H. The Day AI wins the Nobel Prize. 1st Think X Symposium Theme: Smart Healthcare for Smart Nation. Infuse Theatrette at Fusionopolis, Singapore. 2015.04.09.
- Asai Y, Abe T, Kitano H. PhysioDesigner: A versatile platform for multilevel modeling of physiological systems network. BioNetVisA 2015 workshop. National University of Singapore (NUS), Singapore. 2015.11.25
[Poster presentations]
- 浅井義之. 安部武志. 李俐. 北野宏明. 多階層生体機能モデリングプラットフォームによるモルフォロジー・時系列データの統合. 第54回日本生体医工学会大会 . 名古屋国際会議場 (名古屋市). 2015.05.07. [Asai Y, Abe T, Li L, Kitano H. Integration of morphological and timeseries data into mathematical models on multilevel modeling platform. The 54th Annual Conference of Japanese Society for Medical and Biological Engineering. Nagoya Congress Center, 2015.05.07]
- Wagatsuma H, Asai Y, Nomura T. Infrastructure for Neuroscience Ontologies Bridging Between Experimental Data in 3D Atlas, Computational Modeling and Analytical Tools: A basement Approach of DynamicBrain Platform (DB-PF) toward Inter-Platform Coordination in the J-Node. Neuroinformatics 2015. Cairns, Australia. 2015.08.21]
- Ishihara A, Ikeno H, Wagatsuma H, Inagaki K, Usui S, Satoh S, Yamazaki T, Kannon T, Yamaguchi Y, Okumura Y, Kamiyama Y, Asai Y, Hirata Y. Development of an on-line simulation platform for neuroscience research. Neuroinformatics 2015. Cairns, Australia. 2015.08.20]
- Hsin KY, Matsuoka Y, Watanabe T, Kawaoka Y, Kitano H. In-silico approach in assessing anti-influenza agents and their targets using comprehensive pathway map (FluMap). ICSB 2015. Biopolis, Singapore. 2015.11.23.]
- Hsin K-Y, Matsuoka Y, SamikGhosh, Kitano H. Application of machine leaning approaches in drug target identification and network pharmacology aDVANCES IN SYSTEMS AND SYNTHETIC BIOLOGY. Evry, France. 2016.03.21]
- Hsin K-Y, Matsuoka Y, Ghosh S, Kitano H. Combining machine learning systems and multiple docking simulations for network pharmacology. OpenEye Scientific JCUP VI. Ohtemachi SunSky Room, Tokyo. 2015.06.04.]
- Asai Y, Kido Y, Kamiyoshi K, Nomura T, Kitano H. An Ontology-Based Model Search Extended to Deal with Accessibility Issues of Neuroinformatics Metadata with Multidirectional Attributes INCF Japan Node International Workshop: Advances in Neuroinformatics 2015 (AINI 2015). Univ, of Tokyo, JAPAN. 2015.11.25]
[Seminars]
- 浅井義之. 北野宏明. PhysioDesignerプロジェクトにおける可視化ツール. ブレインアトラス・アイデアソン RIKEN (埼玉県和光市) 2015.07.16. [Asai Y, Kitano H. Visualization tools developed in PhysioDesigner project. Brainatlas Ideathon, RIKEN. Wako, Saitama, Japan. 2015.07.16.
- 浅井義之. Life: Dynamical Systems and Modeling. 沖縄高等専門学校 (沖縄県) 2015.07.13. [Asai Y. Life: Dynamical Systems and Modeling.. Okinawa National College of Technology, 2015.07.13.]
- Asai Y, Kitano H. Multilevel modeling and simulation with PhysioDesigner as a platform for collaborative development of models and simulation studies. OIST, Okinawa. 2015.04.09.
- Asai Y, Abe T, Kitano H. Multilevel modeling of physiological system using PhysioDesigner. University of Torino, Turin, Italy. 2015.08.24.
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 Seminar "Identification and design of cancer immunotherapy agents: structure-based drug design of indoleamine 2,3-dioxygenase (IDO) and tryptophan 2,3-dioxygenase (TDO) inhibitors"
- Date: March 2nd, 2016
- Venue: OIST Campus C210
- Speaker: Professor Su-Ying Wu
Investigator, National Health Research Institutes, TaiwanInstitute of Biotechnology and Pharmaceutical Research
7. Other
- PhysioDesigner v1.4 (2016.2.1.), v1.3 (2015.7.14.)
- Flint v1.4 (2016.2.1.), v1.3.1 (2015.7.24.), v1.3 (2015.7.14.)
- PHPlotter v1.0
- ISD Conv v1.1
- DBPF Client v1.1
- PHDB Client v1.1
- PH Database



