FY2011 Annual Report
Open Biology Unit
Professor Hiroaki Kitano

Abstract
Our unit aims to develop a novel software platform for systems biology and systems drug discovery that may fundamentally transform the way we study these fields.
Modern biology and medicine are fundamentally data- and evidence-driven, and researchers are fighting against the vastness and complexity of the data and the scattered nature of our knowledge. In order to obtain in-depth insights that may lead to new biological discoveries and medical implications, one must be able to properly access this data and knowledge and then integrate, analyse, and link them to practical solutions. This requires a new approach in science in the sense that it has to be open-ended, evolvable, community-based, and computationally supported.
Our unit is developing a methodology and software platform for open biology that entails the above features. This includes the Payao community-based pathway curation system, PhysioDesigner physiological modeling tool, drug-molecular network interaction prediction software, and a series of software packages that will be interoperable to The Garuda Platform. The Garuda Platform is a global alliance that we initiated to create a consistent, integrated user experience - a one-stop service style software platform. Such a platform will enable researchers to explore system-level characteristics of biological systems more effectively than before. At the same time, we are exploring a novel drug discovery approach based on computational systems biology, theory of biological robustness, and a concept of long-tail drugs.
1. Staff
- Dr. Yoshiyuki Asai, Group Leader
- Dr. Kun-Yi Hsin, Researcher
- Takeshi Abe, Technical Staff
- Kyota Kamioshi, Technical Staff
- Midori Tanahara, Research Administrator
2. Collaborations
- Theme: Software platform for systems biology
- Type of collaboration: Research Collaboration (no financial transaction)
- Researchers:
- Dr. Samik Ghosh, Systems Biology Institute
- Ms. Yukiko Matsuoka, Systems Biology Institute
- Theme: Integration of Manchester text mining system to Payao
- Type of collaboration: Research Collaboration (no financial transaction)
- Researchers:
- Professor Sophia Ananiadou, Manchester University
- Theme: Open platform for multi-level modeling
- Type of collaboration: Joint research
- Researchers:
- Professor Taishin Nomura, Osaka University
- Professor Hideki Oka, RIKEN
- Professor Yoshihisa Kurachi, Osaka University
- Professor Kun-ichi Hagihara, Osaka University
- Dr. Masao Okita, Osaka University
3. Activities and Findings
3.1 Garuda Platform

One of the major focus of the unit is to launch and lead the Garuda Alliance that is designed to remedy various interoperability issues in systems biology and biomedical software tools and data resources. It aims at providing a coherent and comprehensive software, data, and knowledge platform for systems biology and biomedical research to maximize efficacy of software and computational oriented research while significantly benefitting end-users (typically bench biologists, bioinformatics person at the user site, and those who are in related industrial sectors).
One of the Garuda Alliance’s intentions is to provide a novel open innovation platform that may dramatically improves R&D productivity of basic biology research and pharmaceutical industry. We strived to accomplish these objectives by providing mechanisms for global distribution of computational, software, and knowledge base resources as a one-stop service portal, unified application program interface (Garuda Core API), consistent user experiences, and solid user supports. In FY2011, the prototype of the fundamental platform has been developed, which allows different systems biology tools, databases and services to interoperate seamlessly through open, standardized interfaces enabling researchers to leverage their unique features in a research pipeline.
A single portal site was opened in September 2011 for preview, that will enable users to download software and subscribe to web-based resources with easy-to-access interface as well as enforce a flexible but consistent licensing scheme that enables both academic and commercial entities to join the initiative (Fig. 1). System development companies such as Mitsui Knowledge Industry (MKI) have agreed to modify their commercial software packages to comply with Garuda API. Several companies that provide major scientific computing packages and experimental materials have expressed interest and in discussion for their involvement, such companies involves Mathcore / Wolfram Research, etc. There is a major pharmaceutical company already decided to introduce this system as a part of research platform (name cannot be disclosed at this moment).
Fig. 1 Website of Garuda Alliance opened in 29 September, 2011.
3.2 Development of software for multilevel modeling: PhysioDesigner

PhysioDesigner is an application to support users to build mathematical models of multi-level physiological systems. Models built on PhysioDesigner are written in PHML format, which is an XML based specification taking over from ISML. We have rebranded ISML to PHML in December, 2011. We have released PhysioDesigner 1.0 alpha on January 2012 at physiodesigner.org (Fig. 2).
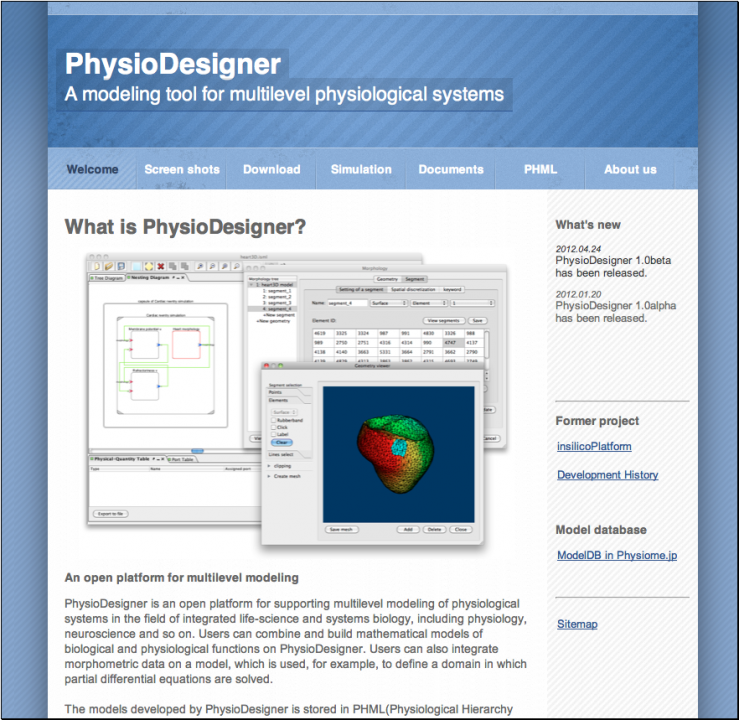
Fig. 2 Snapshot of the official website for PhysioDesigner.
The major achievement on PhysioDesigner development in FY2011 was a completion of renovation of graphical user interface (GUI) based on Java technology (Fig. 3). This development includes reengineering of API calling method from GUI side, based on data serialization technique between two processes. PhysioDesigner has caught up ISIDE’s functions and been developed further mainly in GUI representation and morphology related technology.
The way to deal with morphometric data on PhysioDesigner has been advanced. PhysioDesigner equips a morphology dialog on which users can set geometries of physiological systems and define segments on geometries. If users have morphometric data in formats of VRML, VTK, STK and so on, these data can be uploaded as geometries and integrated into a model on PhysioDesigner. Besides, even if users do not have morphometric data, users can compose a simple morphology using pre-defined primitive shapes such as cone, cuboid, cylinder, sphere and so on. The method to define segments on the morphology editor was also improved. Users can select pieces of mesh interactively by mouse operation and set them as a segment. Segments are used to define initial conditions and boundary conditions for solving partial differential equations. The setup of association between those mathematical expressions and segments (morphology) can be configured on the physical-quantity dialog.
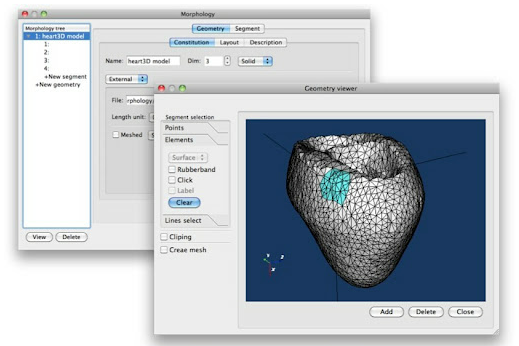
Fig. 3 Snapshots of PhysioDesigner.
3.3 Development of software for simulations of multilevel physiological models: Flint

Flint is a simulator that can parse PHML and SBML. This was called insilicoSim previously. We have rebranded it to “Flint” on January 2012 and released 1.0 alpha in January 2012 (Fig. 4). Flint was reengineered for better simulation performance. The user interface and computation engine were separated clearly as a server-client architecture, which can be applied to the cloud computing system. Flint GUI was re-implemented with Java for making it match up with Garuda platform. The new interface equips some new features. For example, during a simulation, graphs of physical-quantities are updated in time as a simulation progresses.
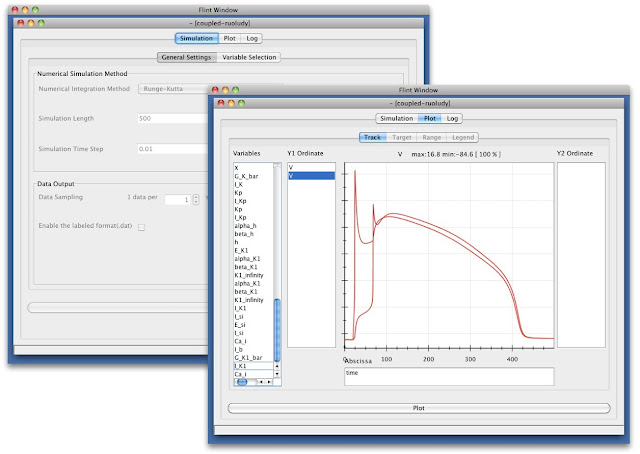
Fig. 4 Flint configuration and simulation windows.
3.4 Development of Flint K3 for cloud and HPC computing

We have begun to develop a compatibility of Flint to high performance computers (HPCs) and clouds. The system Flint K3 (Knit Knowledge Knack) is composed of two servers. One is an interface server (IFS) which is a kind of a front desk of simulation service. IFS receives simulation requests from users, but does not run simulation on it. The tasks are sent to the other server, simulation servers (SSs) located on clouds or HPCs. The SS runs simulations, store simulation results, and return the results to users through IFS.
Flint K3 portal site has been developing on IFS, which manages simulation requests, users, groups, and simulation jobs running on SSs. IFS also equips REST APIs so that applications can access to IFS directly through the APIs. These APIs will be converted as the Garuda API afterwards. Then Garuda enabled applications such as PhysioDesigner and CellDesigner can send simulation jobs directly to Flint K3.
3.5 Development of a web and community based system for pathway sharing: next generation Payao
Payao aims to enable a community to work on the same models simultaneously, insert tags to the specific parts of the model, exchange comments, record the discussions and eventually update the models accurately and concurrently. The current Payao interface was developed by Adobe Flex based on Web 2.0 technology since beginning of the project initiated by SBI. However, for big size pathway maps, the processing speed of Flex application is not enough. This can be a limitation of availability of Payao.
We have reengineered Payao based on JavaScript, with which display of maps and interactive operations can be done quicker than Flex. Including modification of the backbone database schema, Payao JS prototype has reached closer to catch up the current Payao functions (without PathTEXT extension).
In early 2012, SBI side proposed to develop Payao Lite (renamed to iPathways+ lately), which specializes in showing maps with less interactive functions than the current Payao. Instead, as a new feature, we added a function to enable users to embed a map in other web sites, as like google map. A basic feature for embedding maps has been implemented already. This feature has a big potential to expand collaborations with other database sites.
The development of Payao JS and Payao Lite has been promoted jointly by OIST and SBI.
3.6 Development of a computational screening platform for drug discovery and the toxicity prediction
Drugs interact with numbers of molecules within our body. Even when the drug is claimed to have high selectivity, it interacts with molecules that are not identified as a target protein. We aim to create software and knowledge platform that enables us to computationally predict interacting partners of drugs over pathways. Essential facilities include the integration of interaction pathway construction tools and databases, computational docking program, and simulation of perturbation effects over pathways. Through such a comprehensive screening, we can systemically evaluate the potential therapeutic availability and its toxicity of a given drug.
The facilities and resources for above research have been established. A built database compiles a wide range of macro- & small-molecule and Traditional Chinese Medicine (TCM) data. It collects structural and biological information of 8 protein dataset, +80 million small molecules from +130 suppliers and 1 TCM dataset. A very efficient method for unique compound recognition, especially for a large scale database has been developed and is seen to capable of dealing with millions of compounds within a reasonable time. The dataset consisting of unique compounds without redundancy can significantly reduce the run time for performing high-throughput virtual screening.
A plug-in with virtual docking facility built in CellDesigner has been functionally completed and is seen a well integration with CellDesigner (Fig. 5). Users can comprehensively evaluate the binding potential of test compounds against the proteins shown in a pathway map rendered by CellDesigner. Such a screening facility has been implemented as a user-friendly GUI interface and deployed on an in-house computing cluster and OIST HPC, by which non-computer scientists, such as biologist or physicochemist, can quickly setup and perform a high-throughput virtual screening with least data preparation and programming skill required.

Fig. 5 Components of the CellDesigner Plug-in interface with virtual docking facility.
4. Publications
4.1 Journals
- Hiroaki Kitano, Samik Ghosh & Yukiko Matsuoka. Social engineering for virtual 'big science' in systems biology. Nature Chemical Biology. 7, 323–326 (2011) doi:10.1038/nchembio.574. Published online 17 May 2011
- Tiago J.S. Lopes, Martin Schaefer, Jason Shoemaker, Yukiko Matsuoka, Jean-Fred Fontaine, Gabriele Neumann, Miguel A. Andrade-Navarro, Yoshihiro Kawaoka and Hiroaki Kitano. Tissue Specific subnetworks and characteristics of publicly available human protein interaction databases. Bioinformatics. doi: 10.1093/bioinformatics/btr414. Published online 28 July 2011.
- Yoshiyuki Asai and Alessandro E.P. Villa. (2011) Integration and transmission of distributed deterministic neural activity in feed-forward networks. Brain Research. doi:10.1016/j.brainres.2011.10.012
- Samik Ghosh, Yukiko Matsuoka, Yoshiyuki Asai, Kun-Yi Hsin and Hiroaki Kitano. (2011) Software for systems biology: from tools to integrated platforms. Nature Reviews Genetics. Published online Nov. 3, 2011. 12(12), 821-832, Dec. 2011.
- Hidehiko Komine, Yoshiyuki Asai, Takashi Yokoi and Mutsuko Yoshizawa. Non-invasive assessment of arterial stiffness using oscillometric blood pressure measurement. BioMedical Engineering OnLine 2012, 11:6 doi:10.1186/1475-925X-11-6. February 2012
- Carl-Fredrik Tiger, Falko Krause2, Gunnar Cedersund, Robert Palme, Edda Klipp, Stefan Hohmann, Hiroaki Kitano,and Marcus Krantz. A Framework for mapping, visualization and automatic model creation of signal transduction networks. Molecular Systems Biology 8: 578. March 16, 2012.
4.2 Books and other one-time publications
- Yoshiyuki Asai. Isao Nara (supervising editor), Yoshimi Matsuo (editor). Disorder of lower-limb coordination. Physical therapy for Parkinson's disease. Ishiyaku Publisher Inc. pp. 58 - 65. 2011/05 (Non OIST credit)
- Kitano, H. Cancer Robustness and Therapy Strategies. Cancer Systems Biology, Bioinformatics and Medicine (Cesario, A.; Marcus, F.; Eds, Springer), Oct 28. 2011.
- 松岡由希子, 浅井義之, Samik Ghosh, 北野宏明. システム創薬とソフトウエア プラットフォーム.遺伝子医学MOOK別冊「最新創薬インフォマティクス活用マニュアル」(奥野恭史編, 株式会社メディカル ドゥ刊), Oct. 31, 2011.
- Samik Ghosh, Yukiko Matsuoka, Yoshiyuki Asai, Hiroaki Kitano, Anshu Bhardwaj, Vinod Scaria, Rohit Vashisht, Anup Shah, Anupam Kumar Mondal, Priti Vishnoi, Kumari Sonal, Akanksha Jain, Priyanka Priyadarshini, Kausik Bhattacharyya, Vikas Kumar, Anurag Passi, Pratibha Sharma, Open Source Drug Discovery Consortium, Samir Brahmachari. Software Platform for Metabolic Network Reconstruction of Mycobacterium tuberculosis. Systems Biology of Tubarculosis, (ed.) McFadden, et al., Springer (2012) – accepted
4.3 Oral and Poster Presentations
- Hideki Oka, Li Li, Tatsuhide Okamoto, Takahito Urai, Takeshi Abe, Yoshiyuki Kido, Yoshiyuki Asai, Taishin Nomura. Finite Element Method Analysis in insilicoIDE. The 50th Annual Conference of Japanese Society for Medical and Biological Engineering. Tokyo Kanda Campus, Tokyo Denki University, Tokyo. Proceedings April 28 – May 1, 2011.
- Yoshiyuki Asai, Hideki Oka, Taishin Nomura and Hiroaki Kitano. Towards a simulation platform for multi-level neuronal networks. The 3rd International Conference on Cognitive Neurodynamics (3rd ICCN). Niseko, Hokkaido, Japan. June 10, 2011.
- Yoshiyuki Asai and Alessandro E.P. Villa. Distributed deterministic temporal information propagated by feedforward neural networks. International conference on artificial neural networks. Espoo, Finland. June 16, 2011
- Yoshiyuki Asai, Yoshiyuki Kido, Hideki Oka, Taishin Nomura, Hiroaki Kitano. A modeling/simulation platform for multi-level physiological functions. First Group Meeting of HD-Physiology Project, Osaka, Japan. June 20, 2011
- Hiroaki Kitano, Yoshiyuki Asai, Kun-Yi Hsin, Sami Ghosh, Yukuko Matsuoka. Towards an integrated platform for biophysical modeling. First Group Meeting of HD-Physiology Project , Osaka, Japan. June 20, 2011
- Hiroaki Kitano. Origin and progress of systems biology. NC IPSJ-BIO Conference. University of the Ryukyus. June 23, 2011
- Yoshiyuki Asai, Hideki Oka, Takeshi Abe, Masao Okita, Ken-ichi Hagihara, Taishin Nomura, Hiroaki Kitano. An Open Platform toward Large-scale Multilevel Modeling and Simulation of Physiological Systems. The 11th IEEE/IPSJ International Symposium on Applications and the Internet (SAINT2011). Munich, Germany. July 19, 2011.
- Yoshiyuki Asai. An open platform for development of mathematical models in the integrated life science. Dipartimento di Matematica, Università di Torino. Turin, Italy. July 25, 2011.
- Tomohiro Okuyama, Masao Okita, Takeshi Abe, Yoshiyuki Asai, Taishin Nomura, Ken-ichi Hagiwara. Improvement in processing speed of an interpreter-type bio-simulator "insilicoSim" by using GPU. Summer United Workshops on Parallel, Distributed and Cooperative Processing (SWoPP kagoshima 2011). Kagoshima Public Access Center, Kagoshima, Japan. Proceedings July 27-29, 2011.
- Yoshiyuki Asai. CellDesigner & PhysioDesigner Tutorial "PhysioDesigner (Basic usage, Modelng with Morphology and SBML/ISML Hybrid Modeling). Grants-in Aid Innovative Area "HD Physiology Project". Hiyoshi campus, Keio University, Kanagawa, Japan. Aug 20, 2011.
- Kang, H.; Ghosh, S.; Suzuki, K.; Matsuoka, Y.; Kitano, H. Integrated experimental and computational analysis for the dynamics of Tamoxifen resistance in MCF-7 cell lines. International Conference on Systems Biology 2011 (ICSB2011). Rosengarten Conference Center, Mannheim, Germany. Aug 29, 2011.
- Ghosh, S.; Matsuoka, Y.; Kitano, H. iPathways- biological pathways on your palm!. International Conference on Systems Biology 2011 (ICSB2011). Rosengarten Conference Center, Mannheim, Germany. Aug 30-31, 2011.
- Kitano, H. Act Beyond Borders. International Conference on Systems Biology 2011 (ICSB2011), Sysbio Party Night. Rosengarten Conference Center, Mannheim, Germany. Aug 31, 2011
- Kitano, H. How networks can be used to find diagnostic and therapeutic candidates. International Conference on Systems Biology 2011 (ICSB2011) Workshop4: Systems biology to personalize medication and to find novel drug candidates. University of Heidelberg, Germany. Sept 1, 2011.
- Kitano, H. Garuda Vision and Goals. Garuda Five Workshop. University of Heidelberg, Heidelberg, Germany. Sept 1, 2011.
- Takeshi Abe, Yoshiyiki Asai. Demonstration of ISSim as a partner of Garuda alliance. Garuda Five Workshop. University of Heidelberg, Heidelberg, Germany. Sept 1, 2011.
- Kitano, H. Garuda Goals. Garuda Six Workshop. Pavillon Skip, Luxembourg. Sept 28, 2011.
- Yoshiyiki Asai, Takeshi Abe. Demonstration of Linkage among PhysioDesigner, ISSim and Garuda Plotter. Garuda Six Workshop. Pavillon Skip, Luxembourg. Sept 28-30, 2011.
- Kitano, H. HD-Physiology Project and the Garuda Alliance Software and Resource Platform. The 4th Global COE International Symposium on Physiome and Systems Biology for Integrated Life Sciences and Predictive Medicine, Senri-Hankyu Hotel, Osaka, Nov 21, 2011
- Kitano, H. Systems Drug Design. Lecture at Fudan University, Fudan University, Shanghai, China, Dec. 16, 2011.
- Kitano, H. Systems Drug Design. Lecture at Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai, China, Dec. 16, 2011.
- Kitano, H. HD-Physiology Project Overview. The 1st HD Physiology International Symposium, The University of Tokyo, Jan. 20, 2012.
- Yoshiyuki Asai. Open platform PhysioDesigner for multilevel modeling, simulation and model sharing. The 1st HD Physiology International Symposium: Integrative Multi-level Systems Biology for In Silico Cardiology and Pharmacokinetics. Tokyo, Japan. January 20, 2012.
- Kitano, H. Garuda Overview. Garuda Seven Workshop, OIST, Okinawa. Feb. 21, 2012.
- Yoshiyuki Asai. Demonstration of multilevel modeling and simulation on PhysioDesigner. Garuda Seven Workshop. OIST, Okinawa, Japan. Feb. 21, 2012.
- Kitano, H. Systems biomedicine and their computational platforms. HGM2012, Sydney Convention & Exhibition Centre, March 14, 2012. (invited)
- Kitano, H. Systems biomedicine and their computational platforms. Seminar at IMCS, Institute of Molecular and Cell Biology, Singapore, March 15, 2012. (invited)
5. Intellectual Property Rights and Other Specific Achievements
- JP2010-130513 (PCT/JP2011/058229)
- JP2010-224308 (PCT/JP2011/058323)
6. Meetings and Events
6.1 Dynamic Brain Workshop
- Date: December 17, 2011
- Venue: OIST Campus Lab 1
- Organizers: Dr. Yoko Yamaguchi (RIKEN Brain Center)
- OIST hosts: Dr. Yoshiyuki Asai, Professor Hiroaki Kitano (Open Biology Unit) and Professor Kenji Doya (Neural Computation Unit)
- Speakers:
- Dr. Shiro Usui (RIKEN Brain Center)
- Prof. Hideki Oka (Osaka Univ.)
- Dr. Hiroaki Wagatsuma (Kyutech)
- Dr. Yoshiyuki Asai (OIST)
- Dr. Hiroaki Mozuhara (Kyoto Univ.)
6.2 Garuda Seven Workshop
- Date: February 22-25, 2012
- Venue: OIST Seaside House
- Co-organizers: OIST, Systems Biology Institute, University of Tokyo, Manchester University
- Speakers:
- Dr. Huaiyu Mi (University of Southern California)
- Dr. Natalia Polouliakh (Sony CSL)
- Dr. Akira Funahashi (Keio University)
- Dr. Venakta Satagopam (EMBL)
- Dr. Richard Adams (University of Edinburgh)
- Dr. Reinhard Schneider (EMBL)
- Dr. Samik Ghosh (Systems biology Institute)
- Prof. Hiroaki Kitano (OIST)
- Ms. Yukiko Matsuoka (Systems Biology Institute)
- Prof. Akihiro Hisaka (University of Tokyo Hospital)
- Dr. Takao Shimayoshi (ASTEM, Kyoto)
- Prof. Kenichi Aisaki (NIHS)
- Yoshiyuki Asai (OIST)

6.3 Seminar
- Date: March 28 , 2012
- Venue: OIST Campus Lab1
- Speaker: Prof. Akira Amano (Ritsumeikan University)
- Title: Software support for Heart Model Simulation
6.4 Seminar
- Date: March 28 , 2012
- Venue: OIST Campus Lab1
- Speaker: Dr. Takao Shimayoshi (ASTEM, Kyoto)
- Title: Formal Representation Techniques for Cell Simulation
7. Other
Nothing to report.



