FY2014 Annual Report
Integrated Open Systems Unit
Professor (Adjunct) Hiroaki Kitano
Abstract
Open Biology Unit aims to develop a novel software platform for systems biology and systems drug discovery that may fundamentally transform the way we study these fields.
Modern biology and medicine are fundamentally data- and evidence-driven, and researchers are fighting against the vastness and complexity of the data and the scattered nature of our knowledge. In order to obtain in-depth insights that may lead to new biological discoveries and medical implications, one must be able to properly access this data and knowledge and then integrate, analyse, and link them to practical solutions. This requires a new approach in science in the sense that it has to be open-ended, evolvable, community-based, and computationally supported.
Our unit is developing a methodology and software platform for open biology that entails the above features. This includes the Payao community-based pathway curation system, PhysioDesigner physiological modeling tool, drug-molecular network interaction prediction software, and a series of software packages that will be interoperable to The Garuda Platform. The Garuda Platform is a global alliance that we initiated to create a consistent, integrated user experience - a one-stop service style software platform. Such a platform will enable researchers to explore system-level characteristics of biological systems more effectively than before. At the same time, we are exploring a novel drug discovery approach based on computational systems biology, theory of biological robustness, and a concept of long-tail drugs.
Our unit started the Open Energy System Project with SONY CSL and Okisoukou, funded by Okinawa Prefecture. In this project we ‘ll installed PVs and battery System in faculty houses in OIST campus and each house connects DC power line and communication line. We are developing the distributed energy control system and exploring a novel energy exchange algorithm based on real data and simulation. Under this project we finished to install PVs and battery System in 19 faculty houses in OIST campus and each house connects DC power line and communication line to exchange energy. We are collecting energy consumption, production, and exchange data from this DCOES. We evaluate the DCOES performance and contribute to enhance the DCOES performance by analyzing data and running simulation.
1. Staff
- Prof. Hiroaki Kitano, PI
- Dr. Yoshiyuki Asai, Group Leader
- Dr. Kun-Yi Hsin, Staff Scientist
- Takeshi Abe, Technician
- Kyota Kamiyoshi, Technician
- Kuwae Ken, Technician
- Taichiro Sakagami, Technician
- Miwako Nishimura, Technician
- Kenichiro Arakaki, Technician
- Midori Tanahara, Research Administrator
2. Collaborations
- Theme: Software platform for systems biology
- Type of collaboration: Research Collaboration
- Researchers:
- Dr. Samik Ghosh, Systems Biology Institute
- Ms. Yukiko Matsuoka, Systems Biology Institute
- Theme: Integration of Manchester text mining system to Payao
- Type of collaboration: Research Collaboration
- Researchers:
- Professor Sophia Ananiadou, Manchester University
- Theme: Open platform for multi-level modeling and simulation
- Type of collaboration: Joint research
- Researchers:
- Professor Nomura Taishin, Osaka University
- Professor Hideki Oka, RIKEN
- Professor Yoshihisa Kurachi, Osaka University
- Prrofessor Ken-ichi Hagihara, Osaka University
- Dr. Masao Okita, Osaka University
- Professor Akira Amano, Ritsumeikan University
- Dr. Fumiyoshi Yamashita, Kyoto University
- Theme: Modeling and database with dynamic brain platform
- Type of collaboration: Joint research
- Researchers:
- Professor Hiroaki Wagatsuma, Kyusyu Institute of Technology
- Professor Yoko Yamaguchi, RIKEN
- Theme: Neurosignal analysis
- Type of collaboration: Joint research
- Researchers:
- Professor Aleesandro E.P. Villa, University of Lausanne
- Professor Alessandra Lintas, University of Fribourg
- Theme: 4D Visualization of simulation results
- Type of collaboration: Joint research
- Researchers:
- Dr. Ryo Haraguchi, National Cerebral and Cardiovascular Center
- Dr. Takashi Ashihara, Shiga University
- Dr. Naohisa Sakamoto, Kyoto University
- Theme: Cloud supporting simulation service; Flint K3
- Type of collaboration: Joint research
- Researchers:
- Dr. Nobukazu Yoshioka, National Institute of Informatics
- Dr. Shigetoshi Yokoyama, National Institute of Informatics
- Dr. Masaru Nagaku, National Institute of Informatics
- Professor Ken-ichi Hagihara, Osaka University
- Dr. Masao Okita, Osaka University
- Theme: Discovery of anti-influenza agents and drug
- Type of collaboration: Joint research
- Researchers:
- Ms. Yukiko Matsuoka, Systems Biology Institute
- Dr. Tokiko Watanabe. The Institute of Medical Science, The University Of Tokyo
- Professor Yoshihiro Kawaoka. The Institute of Medical Science, The University Of Tokyo, Japan
- Theme: Open Energy System Project
- Type of collaboration: Joint research
- Researchers:
- Sony CSL researchers
3. Activities and Findings
3.1 Development of software for multilevel modeling: PhysioDesigner
PhysioDesigner is a software that supports to create computable models of physiological systems with spatiotemporal multiple levels. Models built on PhysioDesigner are written in PHML format, which is an XML based specification taking over from ISML. We have rebranded ISML to PHML in December, 2011. In FY2014, we have released four versions of PhysioDesigner, i.e. ver. 1.0 (August, 2014), 1.1 (January, 2015), and 1.1.1 (January, 2015), 1.2 (March, 2015) at physiodesigner.org.
By the developments conducted in FY2014, users were enabled to build models in morphospace, in which models are represented in 3-dimensional space with morphological information. The morphospace can be used to visualize simulation results in combination with morphological information. Besides, PHZ format was introduced to save models, which is a kind of ZIP archive being able to include timeserise, morphological data and SBML models which are integrated in the PHML model. A PHZ file can include all data required to perform simulations, and model sharing became hander and simpler. Another significant new feature was a plugin function. Using the plugin framework, users can add functions with GUI written in JAVA as they want. And many of improvements on user interface and bug fixes were done.
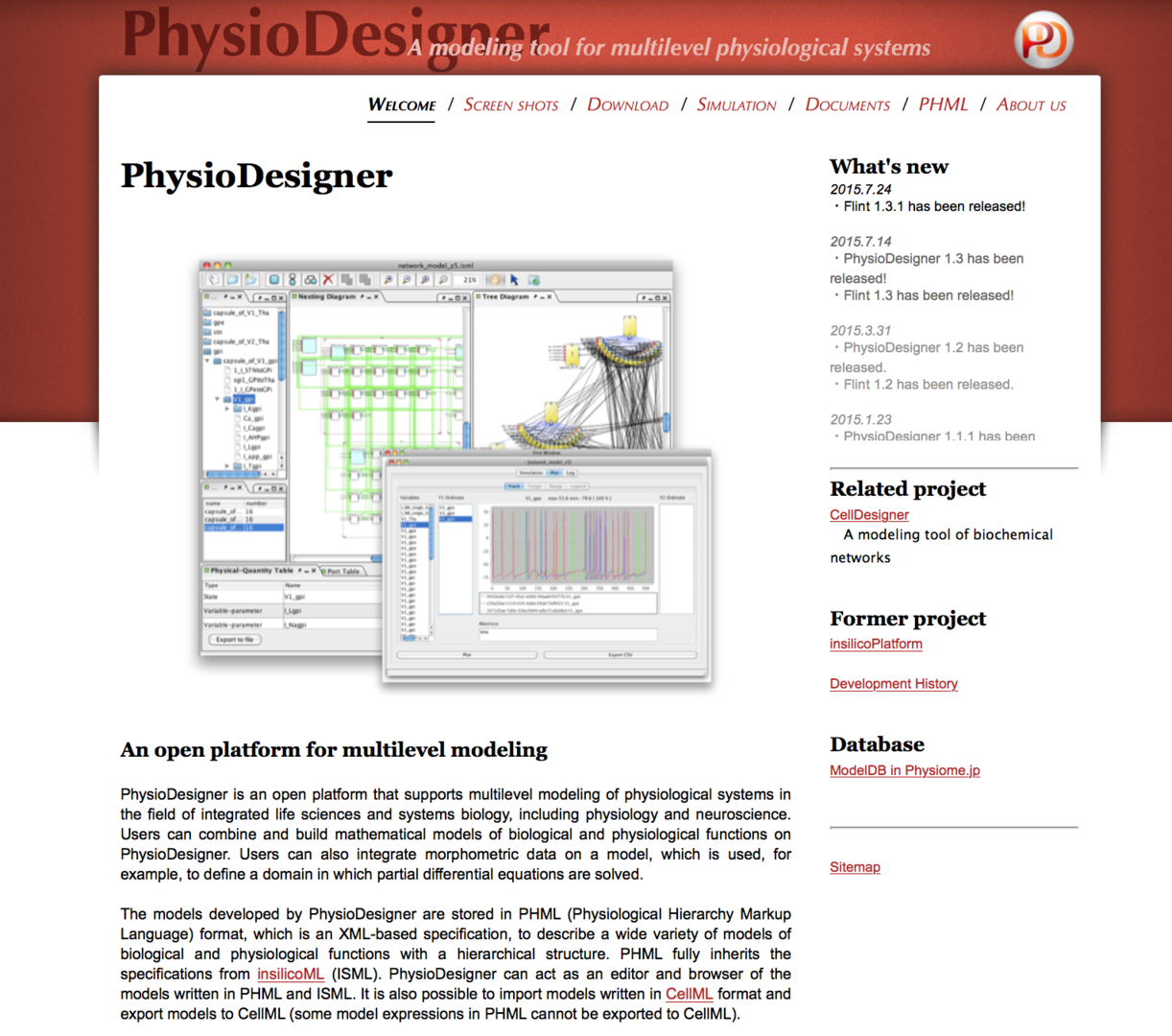
Figure 1: PhysioDesigner official site snapshot
3.2 Development of software for simulations of multilevel physiological models: Flint
Flint is a simulator for a multilevel physiological models described in PHML/SBML. In FY2014, we have released Flint 1.0 in August 2014. Then we have released ver. 1.0.1 in September, 1.02 in October, 1.1 January 2015, and 1.2 in March 2015. Improvements of simulation performance, and improvements of user interface were major development. It was enabled for Flint to handle multiple models at the same time.
3.3 Development of software for visualization of simulation results: PhysioVisualizer
It is possible to create multilevel large scale computable models considering the shape of the physiological systems on PhysioDesigner. To intuitively understand the simulation results of such models, appropriate visualization is necessary. We have been developing PhysioVisualizer, which can visualize such simulation results with the morphological information. In FY2014, the stability and visualizing performance were significantly improved. Besides, a clear-voxel mode was added to visualize simulation data, which can clearly display volume objects even if objects are not well-displayed with the normal rendering method. PhsyioVisualizer is available at Physiome.jp.
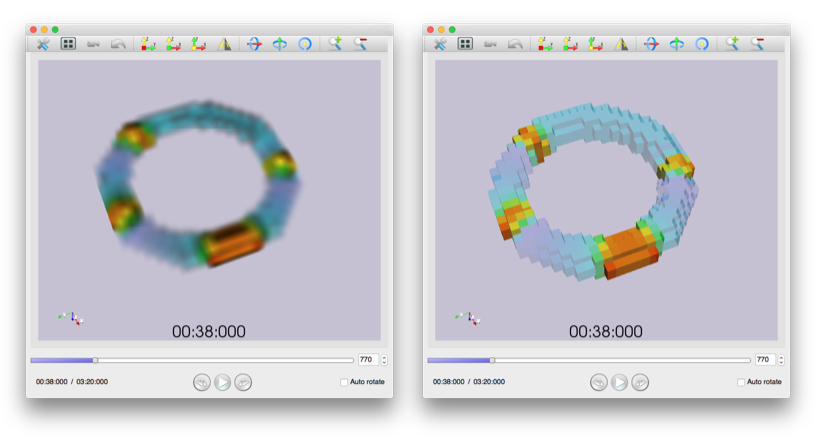
Figure 2: PhysioVisualizer snapshot. Left: the visualization with normal rendering method. Right: with clear-voxel mode.
3.4 Development of Flint K3 for cloud and HPC computing
Since the size of models is getting larger and larger nowadays, to enable Flint to work on high performance computers is demanded. We have been developing a Flint server, called Flint K3, working on computer clouds, so that users of PhysioDesigner can immediately send simulation jobs to high performance computing environment even if users do not have any accesses to high performance computers.
We started to develop K3 with “edubaseCloud” in National Institute of Informatics (NII). But we have migrated Flint K3 to NII’s “Academic Inter Cloude (AIC)”, which is an advanced cloud system than edubaseCloud.
K3 is composed of two servers. One is an interface server (IFS), which receives job requests from users and manage the jobs. Simulation jobs are sent from IFS to simulation servers (SS) in the clouds. To maintenance these servers, we have developed a system state monitoring tool, which can observe the system load and memory usage as line charts.
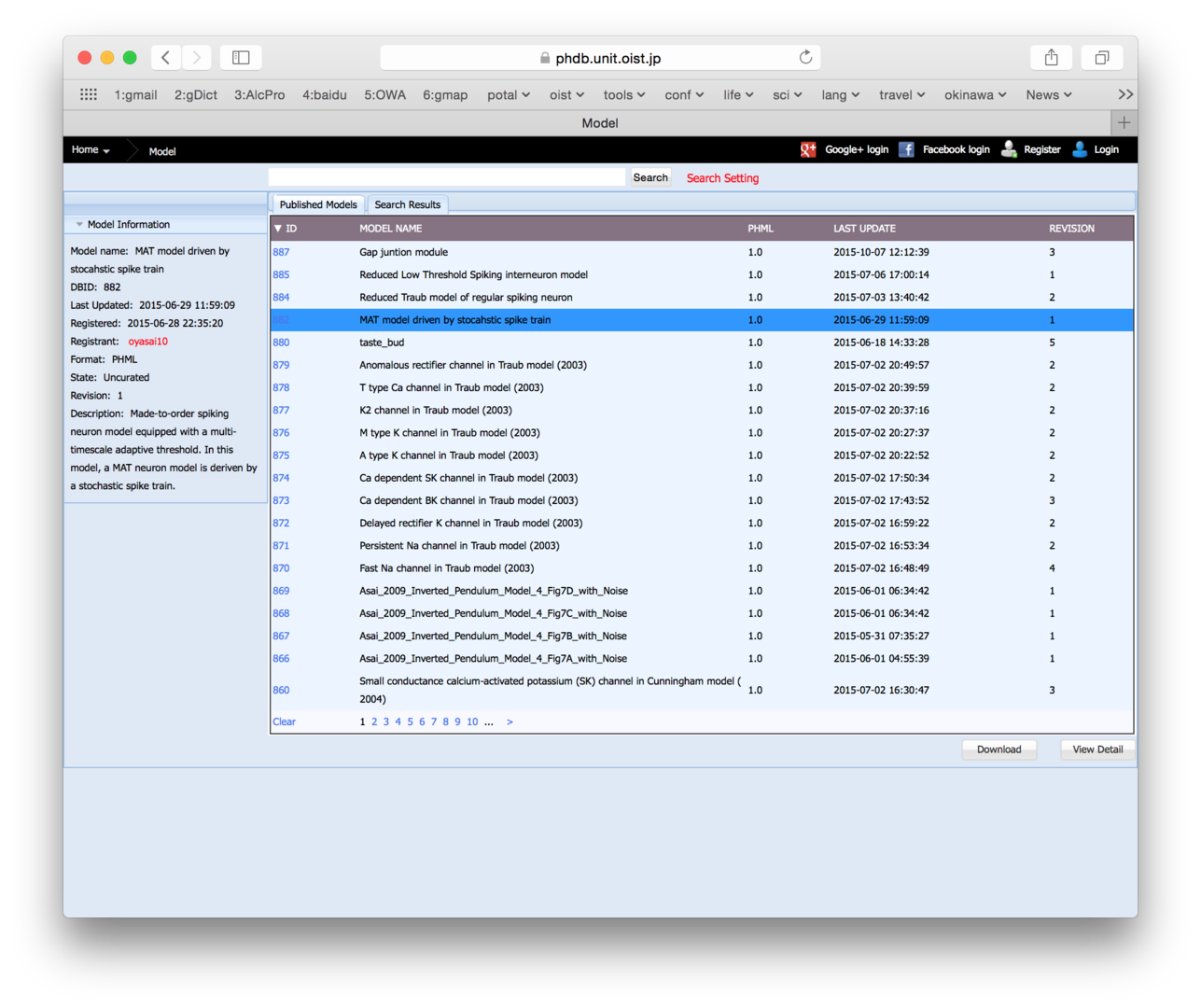
Figure 3: Flint K3 website snapshot
3.5 PH Database
We have renovated databases to store PHML / PHZ models and morphological and timeseries data files at http://physiome.jp. Data management and user management were significantly improved. Major improvements were the followings. The three databases cooperate with each other. A tree-structure of models can be viewed on a web browser. User accounts are managed. Sharing level of the models can be controlled. Update histories of models are logged. Sets of parameter values for a model can be managed. Interoperation with modeling and simulation tools. Interoperation with the Garuda platform.
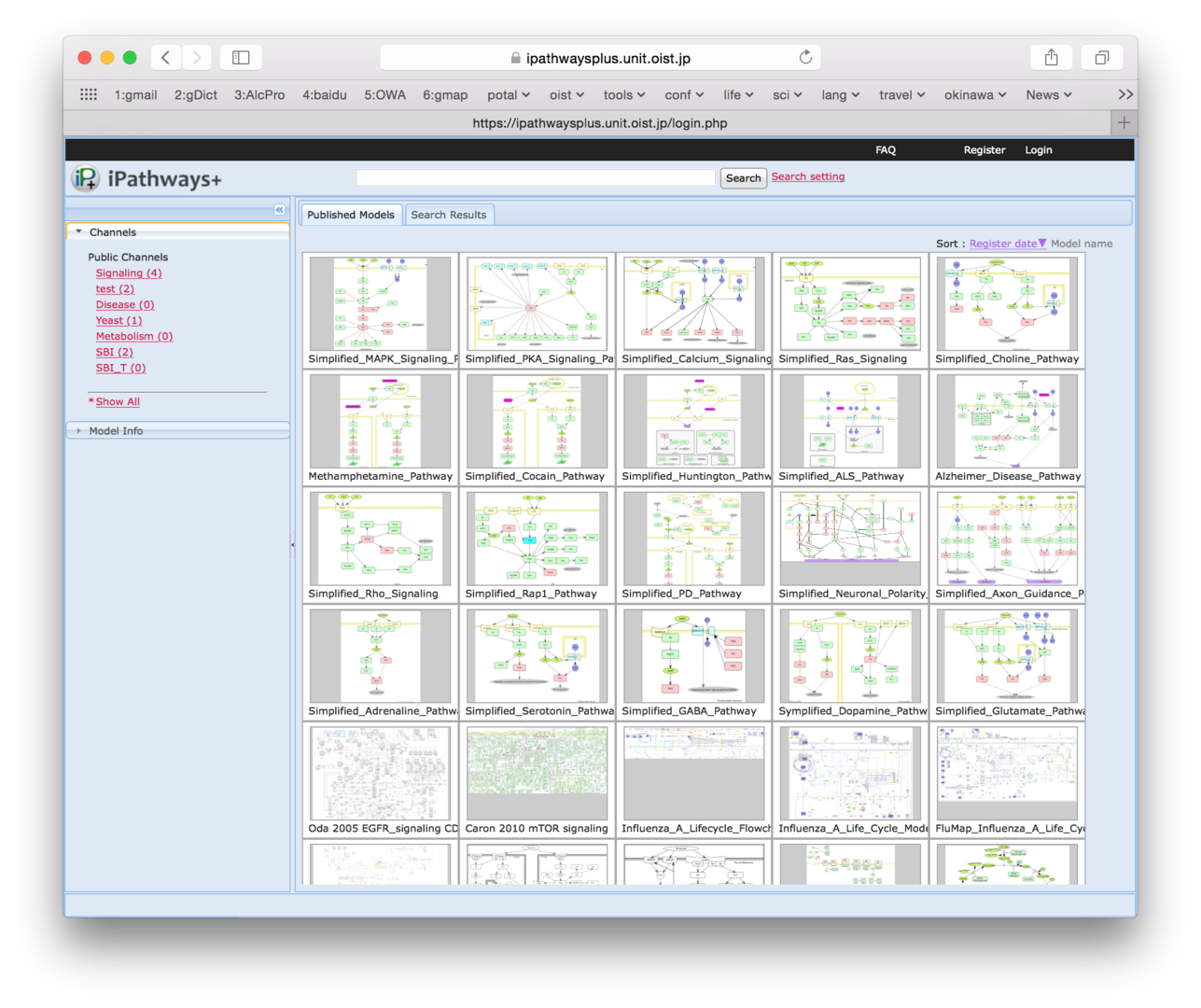
Figure 4: Physiome.jp website snapshot
3.6 Development of predictive system for network pharmacology-based prediction and analysis.
Drugs may interact with multiple molecules in the human body, and such drug action, known as polypharmacology, may be efficacious or deleterious for the treatment of disease. A multi-target strategy has advanced the treatment of diseases. However, poor drug selectivity may increase therapeutic risks and negatively impact drug development because of unintended drug-target interactions. Identification of drug targets is therefore a critical stage of drug development.
With the increased availability of bioinformatics technologies, opportunities for the application of network pharmacology to predict drug effects and toxicity resulting from multi-target interactions are being created. We have developed a novel screening approach which combines two elaborately developed machine learning systems and the use of multiple docking packages to assess binding potentials of a test compound against proteins involved in a complex molecular network. This screening approach enables systematic prediction of network-dependent effects of a test compound (e.g. lead or drug), which accelerates systems pharmacology-based prediction to address drug safety issues. The developed screening systems significantly enhance the prediction accuracy. Using the method, we have screened the compounds targeting the host proteins identified by siRNA screening, and identified those showing efficacy in vitro (illustrated in Figure X). This would contribute in reducing the number of tests for further bioassay. Together with a curated pathway map, the proposed screening approach is able to comprehensively characterize the underlying mechanism of a drug candidate and also to interpret its cascade effects, leading a step change in the prediction of drug efficacy and safety.
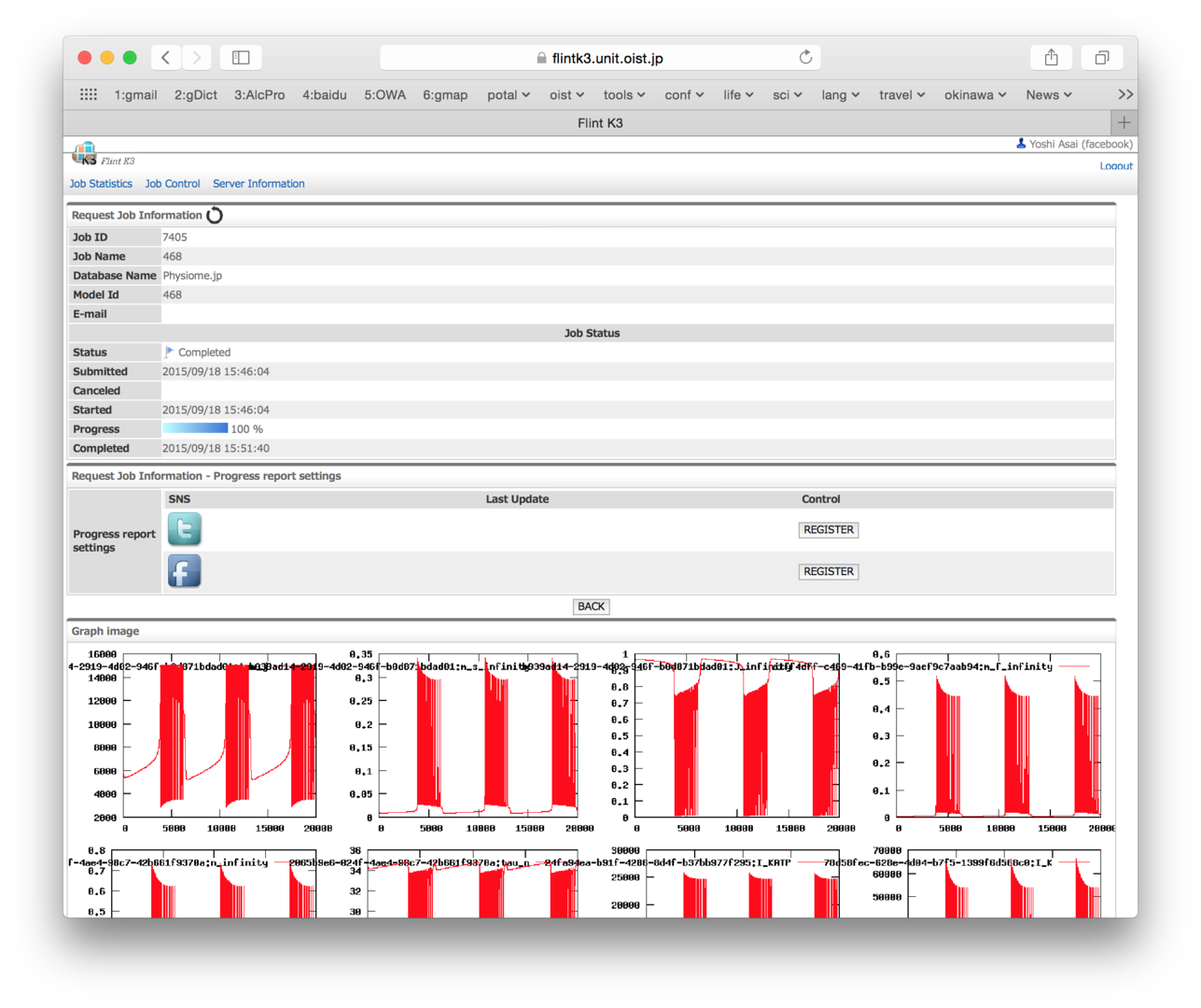
Figure 5: Prediction of a test compound over Influenza A Virus Life Cycle pathway map (FluMap) for discovering anti-influenza agents. FluMap describes the intermolecular network of influenza A virus life cycle. Docking scores are converted into white/red color according to a cutoff value at 5.52 pKd (white: inactive; red: active; gray: not in the test or no results). Results are projected on the pathway map for a comprehensively inspection and investigation.
3.7 Development of a web and community based system for pathway publisher: iPathways+ and Pub2Path
Open Biology Unit and SBI have jointly developed iPathways+ and Pub2Path focusing on the feature of publishing the map models. Both of them do not equip interactive functions. iPathway+ has a function to enable users to embed a map in other web sites, as like google map. This feature has a big potential to expand collaborations with other database sites. iPathways+ is opened at http://ipathways.org/plus/. In the FY2014, a function to categorize and label models was added to help users to find model more easily.
Besides, we have started to develop Pub2Path system, which assists users to implement pathway models which are published on papers. It can store tiny pieces of pathways and sentences described in papers. Utilizing those pieces, users can build a whole pathway map.
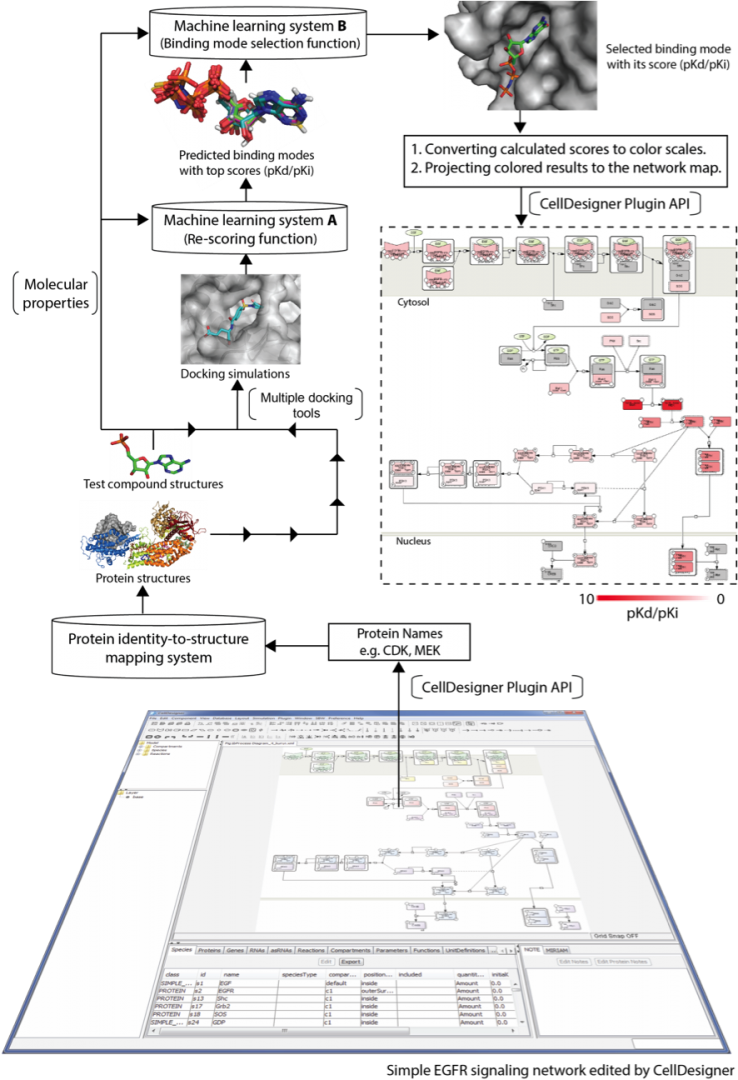
Figure 6: Snapshot of iPathways+
3.8 Open Energy Project
From December 2014, DCOES are operated in 19 faculty houses in OIST campus. DCOES real data in January 2015 show average 97.3 kWh energy generated and supplied to residents. Around 10-15 kWh energy was exchanged per day, and 50 kWh surplus energy were utilized by energy exchange. From summer simulation result, DCOES might utilize around 300 kWh surplus energy by exchanging energy.
4. Publications
4.1 Journals
- P. J. Edmunds, R. Steneck, R. Albright, R. C. Carpenter, A. P. Y. Chui, T.-Y. Fan, S. Harii, H. Kitano, H. Kurihara, L. Legendre, S. Mitarai, S. Muko, Y. Nozawa, J. Padilla-Gamino, N. N. Price, K. Sakai, G. Suzuki, M. J. H. van Oppen, A. Yarid and R. D. Gates. Geographic variation in long-term trajectories of change in coral recruitment: a global-to-local perspective. Marine and Freshwater Research, 66, Feb. 4, 2015
- Kitano, H. Building Cities to Withstand the Worst. Scientific American / Pour La Science, Innovation special issue, 36-37, Jan 2015
- Tokiko Watanabe, Eiryo Kawakami, Jason E. Shoemaker, Tiago J.S. Lopes, Yukiko Matsuoka, Yuriko Tomita, Hiroko Kozuka-Hata, Takeo Gorai, Tomoko Kuwahara, Eiji Takeda, Atsushi Nagata, Ryo Takano, Maki Kiso, Makoto Yamashita, Yuko Sakai-Tagawa, Hiroaki Katsura, Naoki Nonaka, Hiroko Fujii, Ken Fujii, Yukihiko Sugita, Takeshi Noda, Hideo Goto, Satoshi Fukuyama, Shinji Watanabe, Gabriele Neumann, Masaaki Oyama, Hiroaki Kitano, and Yoshihiro Kawaoka. Influenza Virus-Host Interactome Screen as a Platform for Antiviral Drug Development. Cell Host & Microbe, 16, 6, 795-805, Dec.10, 2014
- Asai, Y., T. Abe, H. Oka, M. Okita, K. Hagihara, S. Ghosh, Y. Matsuoka, Y. Kurachi, T. Nomura and H. Kitano (2014). "Versatile Platform for Multilevel Modeling of Physiological Systems: SBML-PHML Hybrid Modeling and Simulation." Advanced Biomedical Engineering (ABE) 3: 50-58.
- 北野宏明, 徳田佳一, 坂上太一郎, 田島茂, 浅井義之, アネッタヴェルツ, 西村美和子, 森田直, 吉村司, 所眞理雄 Open Energy Systems(OES)太陽エネルギー Vol. 40. 5. 2014.12 http://www.jses-solar.jp/wordpress/wp-content/uploads/2014soumokuji.pdf
- Kitano, H./北野宏明. Introduction to Systems Science for Biologist/生物学者のためのシステムサイエンス入門(第6回). QIAGEN eyes, 11, 7-9, Nov. 2014, Review.
- Kitano, H./北野宏明. AI and Information Platform/情報プラットフォームと人工知能の登場. Experimental Medicine/実験医学2014年11月号, 32, 18, 2981-2988, Nov. 1, 2014, Review.
- Asai, Y.; Abe, T.; Matsuoka, Y.; Ghosh, S.; Kitano, H./北野宏明. Software Platform for Systems Biology/システムバイオロジー研究のためのソフトウェア・プラットフォームの動向. Drug Delivery System, 29, 5, 386-396, Nov. 25, 2014, Review.
- Kitano, H./北野宏明. Revolution in Systems Control for Biology/システム制御学のもたらす変革~人間の認知限界を突破するために~. Experimental Medicine/実験医学2015年1月号, 33, 1, 100-108, Jan. 1, 2015, Review.
- 北野宏明. 生物学者のためのシステムサイエンス入門(第5回)QIAGEN eyes Vol. 10 p7-9.
- 北野宏明. 役立つモデルを作るのに必要なこと. 実験医学. VOl 32. 11. 1797-1802. 2014.07.01
- 北野宏明. モデル構築におけるロバストネスとノイズ. 実験医学. Vol 32. 14. 2302-2311. 2014.09.01
- 浅井義之, 野村泰伸, 北野宏明. Physiome.jp におけるモデルデータベースとモデリング・シミュレーションプラットフォームとの連携。計測と制御2014.05
- 北野宏明. システムバイオロジーをオープニング・ゲームに使う~細胞老化研究の事例. 実験医学 Vol32. 8. 1288-1294. 2014.05.01
- 北野宏明. 腫瘍内遺伝的多様性の数理. SURGERY FRONTIER .2014-6. 72-74 (188-190) 2014.06.01
4.2 Books and other one-time publications
- Kitano, H./北野宏明. Starting Systems Biology Ground Up/Dr.北野のゼロから始めるシステムバイオロジー. Tokyo/東京, Yodosha/羊土社, ISBN 978-4-7581-2054-8, Mar. 2015.
4.3 Oral and Poster Presentations
[Conference invited lectures]
- Kitano, H. Garuda Platform: An integrated software solution for data-driven medical sciences. World Health Summit 2014, Federal Foreign Office, Berlin, Germany, Oct. 20, 2014.
- Kitano, H. The day AI wins Nobel Prize. AAAI Workshop on “Beyond the Turing Test”, Hyatt Regency in Austin, Texas, USA, Jan. 25, 2015.
- Kitano, H. Open Energy Systems: Toward a Global Agenda Solution for Sustainability. The Second International Symposium on Open Energy Systems. OIST, Feb. 3, 2015
- Kitano, H. Disruptive Innovation in Computational Platform for Systems Biology. Grant-in-Aid for Scientific Research on Innovative Area “HD Physiology” The Final HD Physiology Symposium, Osaka University Nakanoshima Center, Mar. 6, 2015.
- Kitano, H. The Day AI Wins Nobel Prize: Toward Advanced Intelligence that Transforms the Way We do Biomedical Research. 2015 Hunter Meeting, Crown Plaza Hunter Valley, Lovedale, Australia, Mar. 17, 2015.
- Kitano, H. The Day AI Wins Nobel Prize: Toward Advanced Intelligence that Transforms the Way We do Biomedical Research. MIME-FIT Workshop: Challenges and Future Directions in Systems Medicine and Health ICT, Clayton Campus, Monash University, Australia, Mar. 19, 2015.
- Hsin, KY. Molecular docking simulations in the era of network pharmacology. 42th QSAR conference. Kumamoto Shin-toshin Plaza, Kumamoto, Japan. 2014.11.13
- Kitano, H. Systems Drug Discovery and Software Platform. Unlocking unique. ICSB 2014. Melbourne Convention and Exhibition Centre. 2014.09.15
- Kitano, H. Systems Biology and Applications. ICSB 2014. Melbourne Convention and Exhibition Centre. 2014.09.16
- Kitano, H. (2014). Multiscale disease systems modelling. 7th Noordwijkerhout Symposium on Pharmacokinetics, Pharmacodynamics and Systems Pharmacology: “SYSTEMS PHARMACOLOGY IN DRUG DISCOVERY AND DEVELOPMENT”. NH Conference Centre Leeuwenhorst, Noordwijkerhout, the Netherlands. 2014.04.24
- Kitano, H. (2014). Open Energy System to mitigate global energy issues. Expert Conference on Development of Island's Sustainable Societies (2014). OIST, Okinawa.
- 浅井義之 / Yoshiyuki Asai PhysioDesignerによる生理機能の多階層モデリング / Multilevel modeling of physiological systems using PhysioDesigner 第88回日本薬理学会年会 / The 88th Annual Meeting of The Japanese Pharmacological Society 名古屋 / Nagoya, Japan, 19 March, 2015
- 北野宏明. システム毒性学の戦略とプラットフォーム技術. 第41回日本毒性学会学術年会 シンポジウム:毒性オミクス-遺伝子発現ネットワークを標的とした、治療、毒性、及びそれらの評価の新動向-. 神戸コンベンションセンター2014.07.02
- 浅井義之, 安部武志 , 北野宏明. フィジオームを指向した階層的モデリング/シミュレーション基盤. 公益社団法人 日本薬剤学会第29年会2014.05.22. 大宮ソニックシティビル
- 北野宏明. 多階層生体機能学のさらなる発展. 多階層生体機能学シンポジウム2014.06.14. ナレッジキャピタルコングレコンベンションセンター, 大阪
[Conference oral presentations/seminars]
- Sakagami, T., Nishimura, M. and Kitano, H. Integrated Open Energy Systems. The 2nd Renewable Energy Workshop. Hawaii Foreign-Trade Zone No 9, Hawaii, USA. 2015.02.10
- Yoshiyuki Asai, Hiroaki Kitano, Alessandro E.P. Villa. A framework for simulation of neural network models driven by experimental spike timing data. Neural Coding 2014, 6 Oct -10, 2014, Versailles Cedex, France
- Kun-Yi Hsin. Discovery of anti-influenza agents and drug targets using pathway map (FluMap) and computational approaches. Collaborative projects for infectious disease and immunity – The University Of Tokyo Japan. 2015.03.06
- Asai, Y., Abe, T., Li, L., Oka, H., Kurachi, Y. and Kitano, H. Development of multilevel neural network modeling integrating spike train data on PhysioDesigner. Advances in Neuroinformatics 2014 (AINI2014), RIKEN, Wako, Saitama, Japan 2014.09.25
- Ghosh, S. and Kitano, H. Fly to the future of biology. ISMB 2014. John B. Hynes Memorial Convention Center, Boston, USA. 2014.07.14
- 浅井義之, 北野宏明 PhysioDesignerによる多階層モデリング / Multilevel modeling by PhysioDesigner,「ネットワークが創発する知能研究会」・「データ指向構成マイニングとシミュレーション研究会」合同秋合宿2014 / SIG-EIN & SIG-DOCMAS autumn joint conference 2014, OIST, Okinawa, Japan, 2014年12月16日/ 16/12/2014
- 浅井義之 次世代多階層生体機能シミュレーション基盤構築と実証研究 / Development of the next-generation multilevel modeling and simulation platform and demonstrative research 新学術領域研究「多階層生体機能学」最終成果報告会 / The final HD Physiology meeting 大阪 / Osaka, Japan, 18 March 2015.
- 浅井義之PhysioDesignerチュートリアル. 新学術領域「統合的多階層生体機能学領域の確立とその応用」若手ワークショップ. ハートピア熱海 (静岡県熱海市) 2014.09.21
- 浅井義之PhysioDesigner 1.0 使用上のコツ. 新学術領域「統合的多階層生体機能学領域の確立とその応用」若手ワークショップ. 秋田大学 (秋田県秋田市) 2014.09.21
- 浅井義之PhysioDesigner と Flint の現状. 新学術領域「統合的多階層生体機能学領域の確立とその応用」全体会議. 秋田大学 (秋田県秋田市) 2014.08.05
- 浅井義之, 安部武志, Li, Li, 岡秀樹, 倉智嘉久, 北野宏明. PhysioDesignerによる多階層生体機能モデリングサポート技術の進展第53回日本生体医工学会大会. 仙台国際センター (宮城県仙台市)2014.06.25
- 浅井義之, 安部武志, Li, L., 岡秀樹, 野村泰伸, 北野宏明. Physiome.jpのモデルデータベースならびにツール連携の開発. 第53回日本生体医工学会大会. 仙台国際センター (宮城県仙台市) 2014.06.26
- Kitano, H. (2014.04.22). Frontiers of Systems Biology and Software Platform. L’ORÉAL Research & Innovation, Aulnay-sous-Bois, France.
- Asai, Y. (2014). Scien ce Cafe: Towards Big Science Campus Plaza Kyoto, Kyoto, Japan.
- Asai, Y., T. Abe, L. Li and H. Kitano (2014). The 2nd PhysioDesigner tutorial. Osaka University Nakanoshima center, Osaka, Japan.
[Conference poster presentations]
- Yoshiyuki Asai, Takeshi Abe, Hiroaki Kitano. “PhysioDesigner 1.2,” The Final HD Physiology International Symposium, Osaka, Japan, 4-5 March, 2015,
- Takeshi Abe, Yoshiyuki Asai, Masao Okita, Miho Nagai, Ken-ichi Hagihara, Hiroaki Kitano. “Flint and Flint K3: Simulations of Biophysiological models,” The Final HD Physiology International Symposium, Osaka, Japan, 4-5 March, 2015,
- Kun-Yi Hsin, Yukiko Matsuoka and Hiroaki Kitano. In-silico approach in assessing anti-influenza agents and their targets using comprehensive pathway map (FluMap) 4th Negative Strand Virus-Japan。Raguna Garden Hotel Okinawa 2015.01.19
- Kun-Yi Hsin, Yukiko Matsuoka, Samik Ghosh and Hiroaki Kitano. Combining machine learning systems and multiple docking simulations for network pharmacology. High Definition Physiology symposium, Osaka University Nakanoshima Center, 2015.03.04
- Hsin, KY., Ghosh, S. and Kitano, H. Using molecular simulations for network pharmacology. EuroQSAR 2014-20th European Symposium. St. Petersburg, Russia. 2014.09.04
- Sakagami, T., Nishime, K., Tokuda, Y. and Kitano, H. DC-based microgrid in Okinawa. The Asia Pacific Resilience Innovation Summit and Expo (APRISE2014). Oafu HI USA. 2014.09.16
- Asai, Y., Kuwae, K., Li, L., Kitano, H., Wagatsuma, H. and Yamaguchi, Y. Interoperability between multilevel modeling platform PhysioDesigner and with databases in Physiome.jp and Dynamic Brain Platform through Garuda platform. Neuroinformatics2014. Leiden, Netherlands. 2014.08.26
- Ashizaki, K.; Yamanaka, R.; Ohta, T.; Aburatani, H.; Kitano, H./芦崎晃一; 山中遼
- ; 油谷浩幸; 北野宏明. Virtual Environment for Quick and Practical Use of Galaxy/Galaxyを迅速に活用するための仮想環境の構築. The 37th Annual Meeting of the Molecular Biology Society of Japan/第37回日本分子生物学会年会; Pacifico Yokohama/パシフィコ横浜; Nov. 27, 2014.
5. Intellectual Property Rights and Other Specific Achievements
-
ネットワークモデル統合装置、ネットワークモデル統合システム、ネットワークモデル統合方法、および、プログラム. 北野宏明, 松岡由紀子, Samik Ghosh, OIST, SBI. 特許第5639237号 (特願2013-159876) 2014年10月31日取得 (国内).
6. Meetings and Events
Nothing to report
7. Other
- PhysioDesigner. http://www.physiodesigner.org
- Flint. http://www.physiodesigner.org/simulation/flint/index.html
- PHDB Client. http://www.physiome.jp/tools/phdbclient/.



