FY2013 Annual Report
Open Biology Unit
Professor (Adjunct) Hiroaki Kitano

Abstract
Open Biology Unit aims to develop a novel software platform for systems biology and systems drug discovery that may fundamentally transform the way we study these fields.
Modern biology and medicine are fundamentally data- and evidence-driven, and researchers are fighting against the vastness and complexity of the data and the scattered nature of our knowledge. In order to obtain in-depth insights that may lead to new biological discoveries and medical implications, one must be able to properly access this data and knowledge and then integrate, analyse, and link them to practical solutions. This requires a new approach in science in the sense that it has to be open-ended, evolvable, community-based, and computationally supported.
Our unit is developing a methodology and software platform for open biology that entails the above features. This includes the Payao community-based pathway curation system, PhysioDesigner physiological modeling tool, drug-molecular network interaction prediction software, and a series of software packages that will be interoperable to The Garuda Platform. The Garuda Platform is a global alliance that we initiated to create a consistent, integrated user experience - a one-stop service style software platform. Such a platform will enable researchers to explore system-level characteristics of biological systems more effectively than before. At the same time, we are exploring a novel drug discovery approach based on computational systems biology, theory of biological robustness, and a concept of long-tail drugs.
Out unit started the Open Energy System Project with SONY CSL and Okisoukou, funded by Okinawa Prefecture. In this project we ‘ll installed PVs and battery System in faculty houses in OIST campus and each house connects DC power line and communication line. We are developing the distributed energy control system and exploring a novel energy exchange algorithm based on real data and simulation.
1. Staff
- Prof. Hiroaki Kitano, PI
- Dr. Yoshiyuki Asai, Group Leader
- Dr. Kun-Yi Hsin, Researcher
- Takeshi Abe, Technician
- Kyota Kamiyoshi, Technician
- Kuwae Ken, Technician
- Taichiro Sakagami, Technician
- Miwako Nishimura, Technician
- Midori Tanahara, Research Administrator
2. Collaborations
- Theme: Software platform for systems biology
- Type of collaboration: Research Collaboration
- Researchers:
- Dr. Samik Ghosh, Systems Biology Institute
- Ms. Yukiko Matsuoka, Systems Biology Institute
- Theme: Integration of Manchester text mining system to Payao
- Type of collaboration: Research Collaboration
- Researchers:
- Professor Sophia Ananiadou, Manchester University
- Theme: Open platform for multi-level modeling and simulation
- Type of collaboration: Joint research
- Researchers:
- Professor Nomura Taishin, Osaka University
- Professor Hideki Oka, RIKEN
- Professor Yoshihisa Kurachi, Osaka University
- Prrofessor Ken-ichi Hagihara, Osaka University
- Dr. Masao Okita, Osaka University
- Professor Akira Amano, Ritsumeikan University
- Dr. Fumiyoshi Yamashita, Kyoto University
- Theme: Modeling and database with dynamic brain platform
- Type of collaboration: Joint research
- Researchers:
- Professor Hiroaki Wagatsuma, Kyusyu Institute of Technology
- Professor Yoko Yamaguchi, RIKEN
-
Theme: Neurosignal analysis
-
Type of collaboration: Joint research
-
Researchers:
-
Professor Aleesandro E.P. Villa, University of Lausanne
-
Professor Alessandra Lintas, University of Fribourg
-
-
-
Theme: 4D Visualization of simulation results
-
Type of collaboration: Joint research
-
Researchers:
-
Dr. Ryo Haraguchi, National Cerebral and Cardiovascular Center
-
Dr. Takashi Ashihara, Shiga University
-
Dr. Naohisa Sakamoto, Kyoto University
-
-
-
Theme: Cloud supporting simulation service; Flint K3
-
Type of collaboration: Joint research
-
Researchers:
-
Dr. Nobukazu Yoshioka, National Institute of Informatics
-
Dr. Shigetoshi Yokoyama, National Institute of Informatics
-
Dr. Masaru Nagaku, National Institute of Informatics
-
Professor Ken-ichi Hagihara, Osaka University
-
Dr. Masao Okita, Osaka University
-
-
-
Theme: Open Energy System Projcct
-
Type of collaboration: Joint research
-
Researchers:
-
Sony CSL resaerchers
-
-
3. Activities and Findings
3.1 Development of software for multilevel modeling: PhysioDesigner
PhysioDesigner is a software that supports to create computable models of physiological systems with spatiotemporal multiple levels. Models built on PhysioDesigner are written in PHML format, which is an XML based specification taking over from ISML. We have rebranded ISML to PHML in December, 2011. In FY2013, we have released six versions of PhysioDesigner, i.e. 1.0 beta4 (April, 2013), 1.0 beta5 (July, 2013), and 1.0 beta6 (October, 2013), 1.0 beta6.1 (November, 2013), 1.0 beta7 (January, 2014) and 1.0 RC1 (March, 2014) at physiodesigner.org (Fig. 2).
The major achievements on PhysioDesigner development in FY2013 were development of functions to integrate timeserise and morphological data. Time series data obtained by either experiments or simulations can be integrated in PHML models on PhysioDesigner. Integrated time series can be used as an input to derive the model, or as constraint condition for the dynamics of the model. Besides that, morphological data can be also integrated with time series data to create movie showing the simulation results.
We also defined Physiological Hierarchy Parameter List markup language (PHPL) which is an XML based language describing a set of parameters (values of static-parameter type physical quantities and initial values of state type physical quantities) to store them apart from PHML model itself. Then a PHPL file can be used to set multiple parameter values in a PHML model at once. Sometimes a single model can represent different physiological phenomena by working with different parameter values. In such a case, if it is possible to switch those parameter values from one model to another model at once, it would be convenient. There is also another kind of demand. During a simulation, sometimes we want to change parameter values when a certain event happens. PHPL will help to switch over a model representing a certain physiological function into a model of another kind of function, and to changing a set of parameter values at once in mid-course of a simulation.
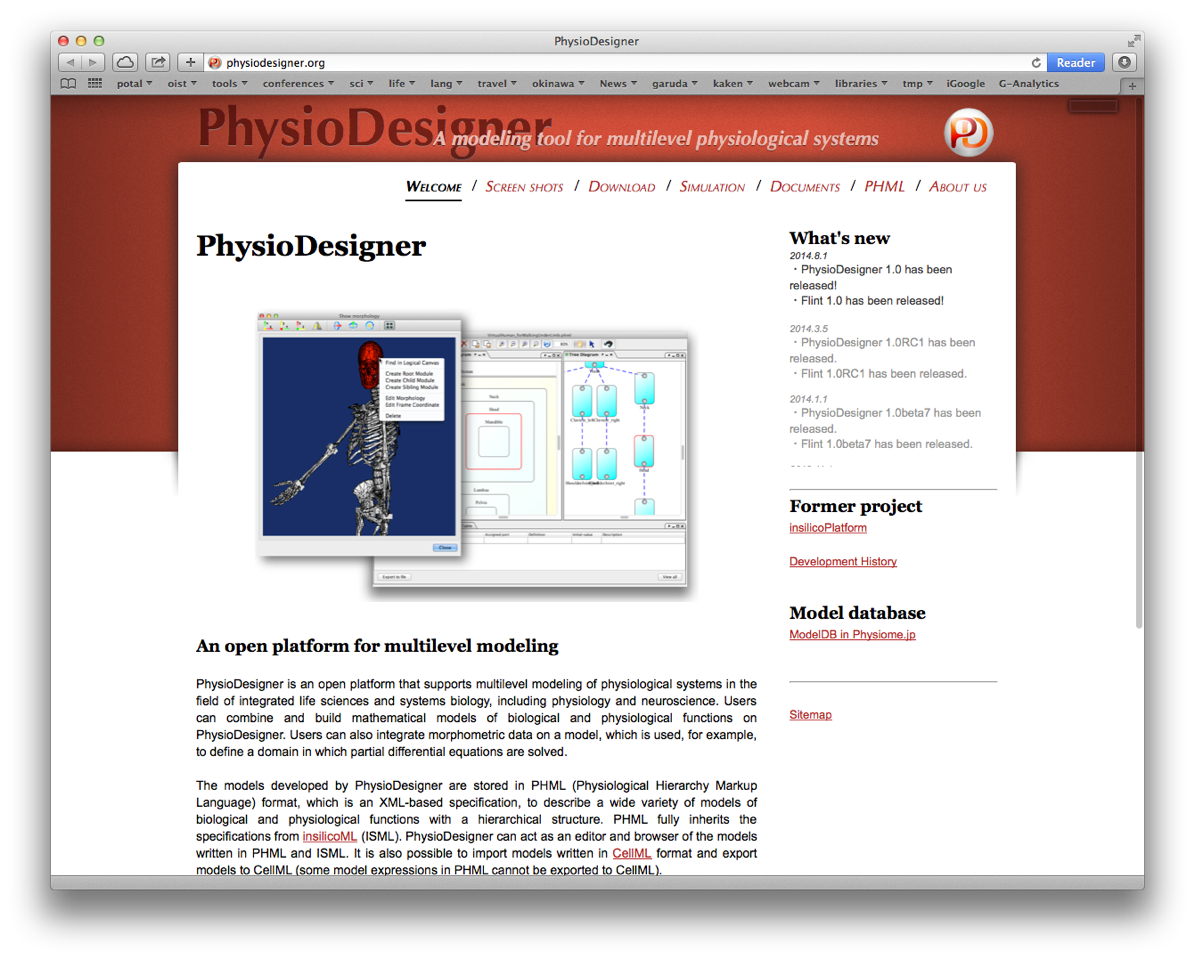
Figure 1: PhysioDesigner official site snapshot
3.2 Development of software for visualization of simulation results: PhysioVisualizer
Using PhysioDesigner, we can create multilevel large scale computable models of physiological systems. Flint can conduct the simulations of those models. We have been developing PhysioVisualizer which can visualize such simulation results and can deal with the morphological object and simulation data with the concept of the layers. Namely it can overlay several layers in one visualization. To superimpose several layers is not simple implementation. A well-prevailed library for visualization called VTK cannot manipulate such cases. Hence we have re-implementing PhysioVisualizer with KVS library. The PhysioVisualizer version 1.0RC has been released on March 2014.
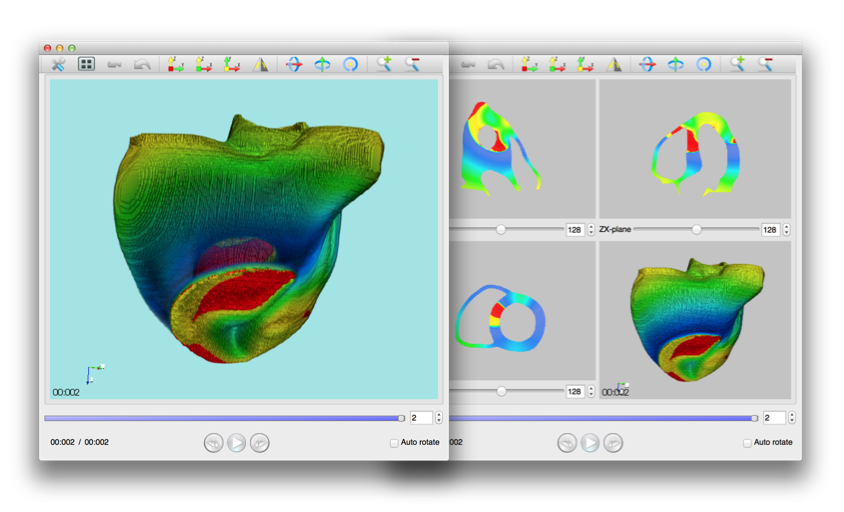 Figure 2: PhysioVisualizer snapshot
Figure 2: PhysioVisualizer snapshot
3.3. Development of software for simulations of multilevel physiological models: Flint
Flint is a simulator aiming to be capable of simulating a multilevel physiological models described in PHML/SBML. At the end of FY2013, we have released Flint 1.0 RC1 for those who interested in running simulations of such models on their desktops. Now it comes with better performance and support of latest PHML features as well as useful functions such as rendering logarithmic scale graphs. We also have enhanced its interoperability with existing desktop and web applications for biology by supporting Garuda API complied with the Garuda alliance (http://www.garuda-alliance.org/).
3.4 Development of Flint K3 for cloud and HPC computing
Since the size of models is getting larger and larger nowadays, to enable Flint to work on high performance computers is demanded. We have been developing a Flint server, called Flint K3, working on computer clouds, so that users of PhysioDesigner can immediately send simulation jobs to high performance computing environment even if users do not have any accesses to high performance computers.
We started to develop K3 with ``edubaseCloud'' (http://edubase.jp/cloud) which is an open source based computer cloud for education of cloud engineering developed in National Institute of Informatics (NII). But now NII provides more advanced system called “Academic Inter Cloude (AIC)”, hence we have prepared to migrate the K3 system to the AIC.
K3 is composed of two servers. One is an interface server (IFS), which receives job requests from users and manage the jobs. Simulation jobs are sent from IFS to simulation servers (SS) in the clouds. SSs evoke a computation program (CP) in every node assigned for the simulation. CPs perform numerical computation of the model in parallel having communications among them.
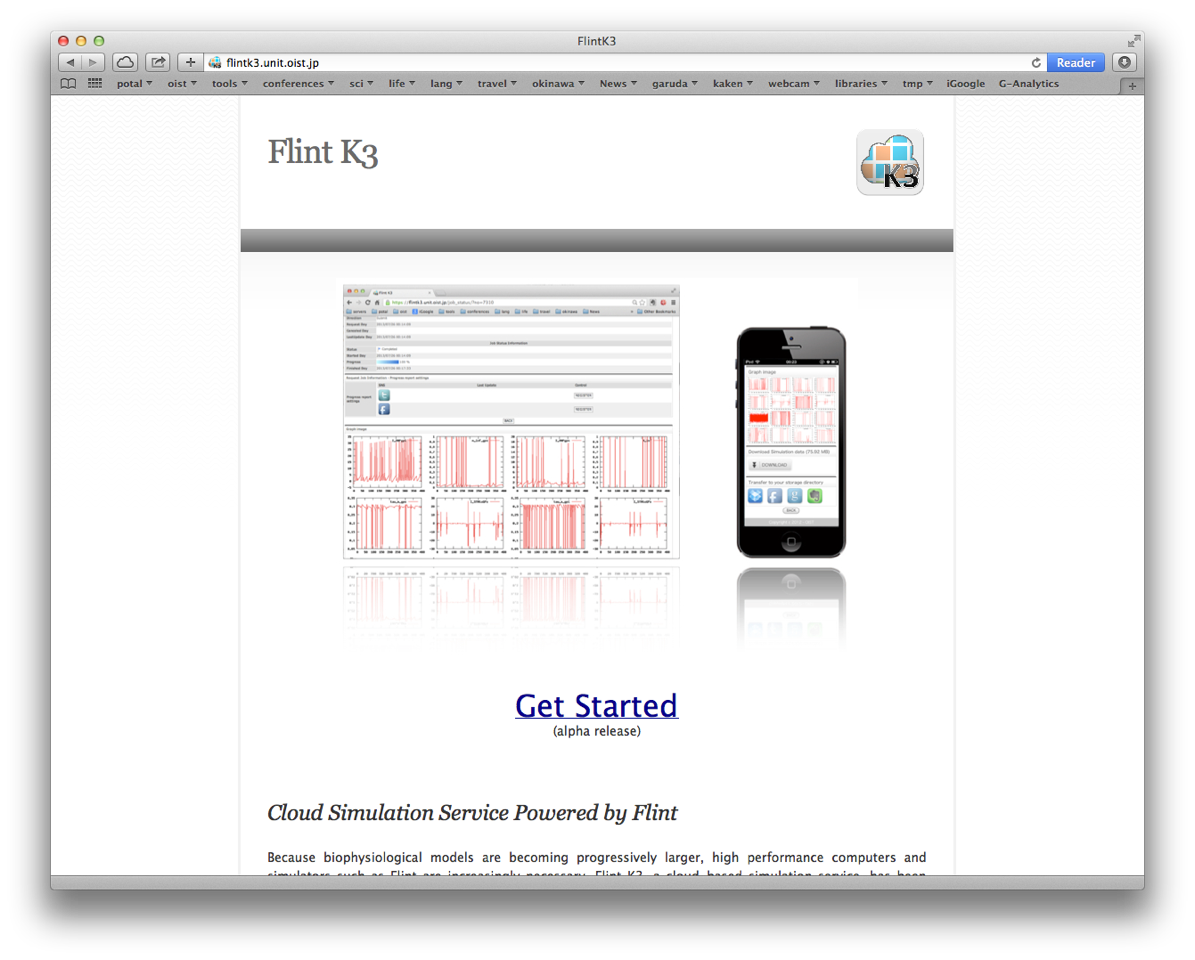
Figure 3: Flint K3 website snapshot
3.5 Porting Flint to The K computer
We have ported Flint to the K computer (http://www.riken.go.jp/en/research/environment/kcomputer/) in order to take advantage of its computation power for simulating large-scale models. This project finished at the end of March 2014. Combining inter-node parallel computation with better scheduling, Flint now calculates both ODE and DDE (Delay Differential Equation) faster, and it became possible to finish a simulation on a large model like heart cells within a feasible time span. The technique developed in this attempt is based on MPI so that it is portable up to recent HPC architecture, i.e., can be applied to porting it to any other HPC facility.
3.6 Physiome.jp
We have renovated the portal site for Physiome opened at http://physiome.jp. This site provides several tools helpful for multilevel modeling, simulation and visualization as well as databases of models, time series data and morphological data.
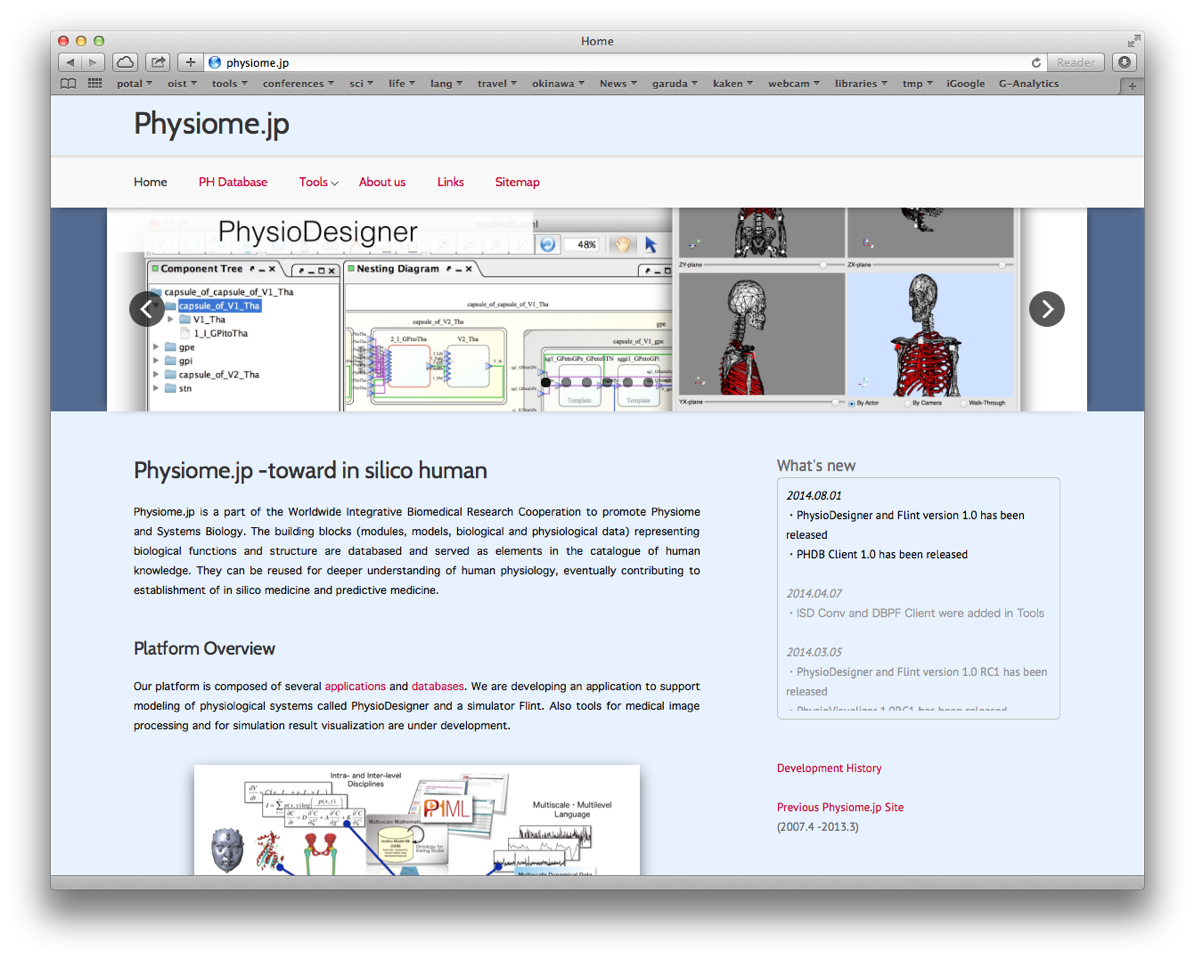
Figure 4: Physiome.jp website snapshot
3.7 Development of a web and community based system for pathway publisher: iPathways+
Open Biology Unit and SBI has jointly developed iPathways+ focusing on the feature of publishing the map models. . iPathways+ does not equip interactive functions, instead, as a new feature, we added a function to enable users to embed a map in other web sites, as like google map. This feature has a big potential to expand collaborations with other database sites. We have released iPathways+ 1.0 at http://ipathways.org/plus/.
Figure 5: iPathways+ website snapshot
3.8 Using molecular simulations for new network pharmacology
Drugs may interact with numerous molecules in the human body. Approximately 35% of known drugs or drug leads present multi-target activity. Even when a drug is claimed to have high selectivity, it probably binds to proteins that are not identified as targets. Such unexpected off-target interactions may result in adverse reactions, which increase therapeutic risks and negatively impact drug development. To comprehensively assess pharmacological effects, systems pharmacology has been developed, in which various bioinformatics resources assessing different structural levels, from molecules to systems are integrated. A well-curated, comprehensive molecular interaction network is the focal point of the systems pharmacological approach. Such a network can reveal causes and effects of protein interactions over signaling networks, metabolic networks, and other related pathways. With a deeply curated network map that describes signaling cascades and interactions among molecules, one can carry out network-based screening to systematically identify target proteins of a given drug candidate and to assess its impact. Thus, network-based screening appears promising for drug repurposing and safety prediction. To accurately predict a drug's effects across molecular networks requires development of high-precision molecular docking simulation systems, and applying them over molecular networks to compute aggregated effects of drugs. We have developed a prediction pipeline for network pharmacology, which consists of a high-precision docking simulation developed in our unit lately (DOI: 10.1371/journal.pone.0083922) and the facility of CellDesigner, illustrated in Figure 6. The prediction pipeline enables drug developers to speed up drug discovery with good accuracy and can be a leading provider of early stage.
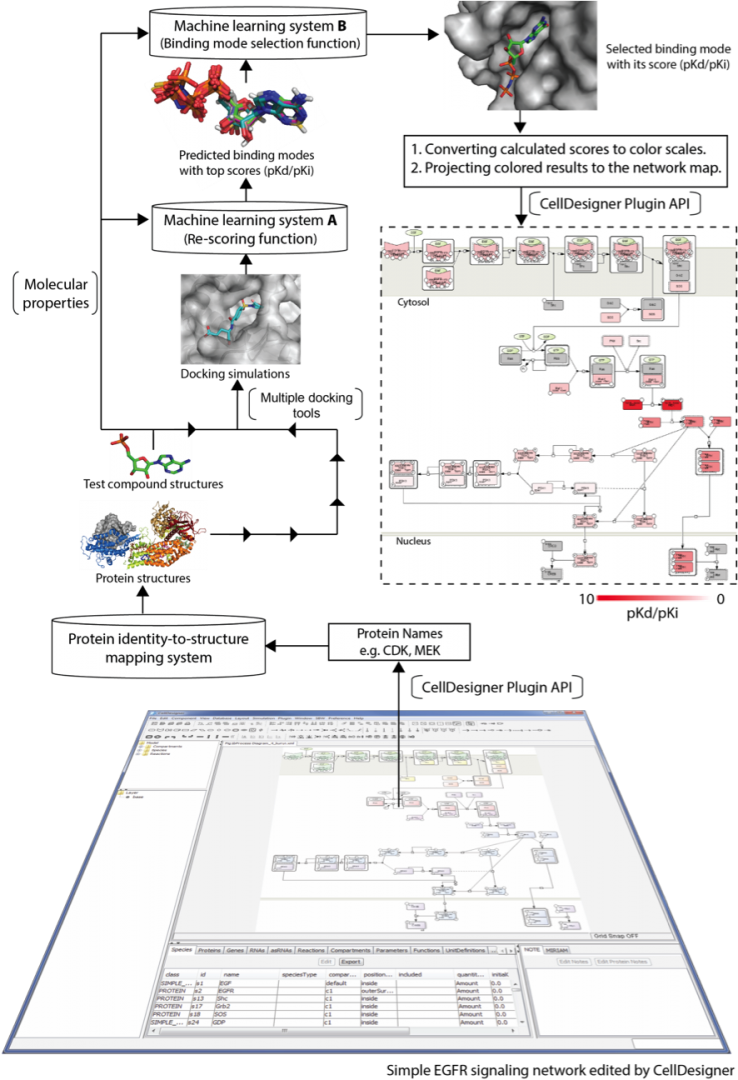
Figure 6: Schematic of the signaling network-based screening pipeline. First, a signaling network is launched by CellDesigner. The identities of proteins involved in the network are retrieved by the CellDesigner plugin API to look up corresponding protein structures in 3D through a protein identity-to-structure mapping system. Second, users submit test compounds for docking simulation. After docking simulation using three docking tools, machine learning system A is then applied to re-score generated binding modes based on features of binding interactions and the test compound's molecular properties, after which, it ranks them. Machine learning system B is subsequently to select a binding mode with the greatest reliability from the three top-score modes. Screening is iterated to assess the next protein until all pathway proteins have been tested. Finally, docking scores are converted into a white-to-red color scale to interpret binding strength, and are projected on the network map for a comprehensive inspection.
3.9 Open Energy System Project
We started to simulate basic energy exchange algorithm based on observed solar irradiation data and the same PV and battery configuration with installed faculty houses. From 16-days simulation output, total 58 kWh energy transfers within 137 transaction are executed between 5 houses and 204 kWh energy transfers with 512 transactions are executed between 17 houses. We ‘ll continue to develop and expand this simulation and explore the novel energy exchange algorithm.
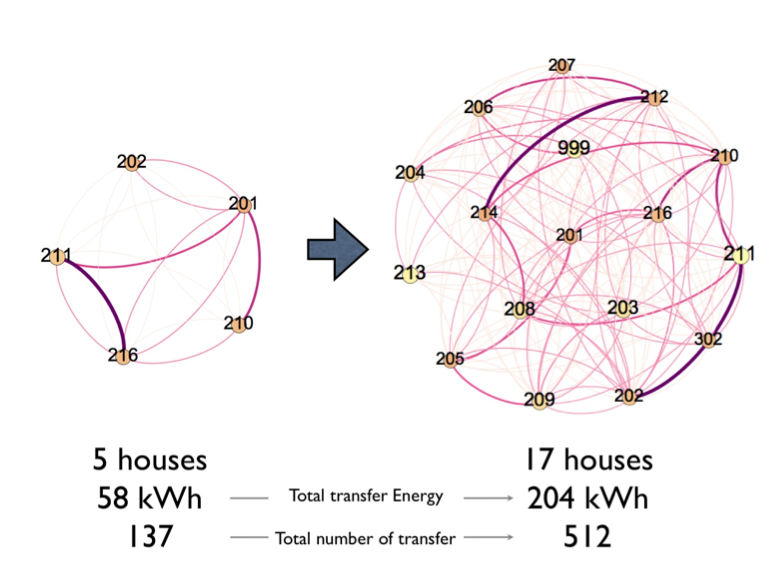
Figure 7: Energy exchange result from our simulation
4. Publications
4.1 Journals
- Asai, Y., et al. (2013). "Learning an Intermittent Control Strategy for Postural Balancing Using an EMG-Based Human-Computer Interface." PLoS One 8(5).
- Fujita, K. A., et al. (2014). "Integrating pathways of Parkinson's disease in a molecular interaction map." Mol Neurobiol 49(1): 88-102.
- Hase, T., et al. (2013). "Harnessing diversity towards the reconstructing of large scale gene regulatory networks." PLoS Comput Biol 9(11): e1003361.
- Hsin, K. Y., et al. (2013). "Combining machine learning systems and multiple docking simulation packages to improve docking prediction reliability for network pharmacology." PLoS One 8(12): e83922.
- Matsuoka, Y., et al. (2013). "A comprehensive map of the influenza A virus replication cycle." BMC Syst Biol 7: 97.
- Yamashita, F., et al. (2013). "Modeling of rifampicin-induced CYP3A4 activation dynamics for the prediction of clinical drug-drug interactions from in vitro data." PLoS One 8(9): e70330.
- Kitano, H. (2013). Introduction to Systems Science for Biologist. QIAGEN eyes. June 2013: 8-12.
- Kitano, H. (2013). Introduction to Systems Science for Biologist. QIAGEN eyes. . September 2013.
- Kitano, H. (2013). Introduction to Systems Biology. Experimental Medicine. 31: 2976-2982.
- Kitano, H. (2013). Introduction to Systems Science for Biologist. QIAGEN eyes. November 2013: 14-16.
- Kitano, H. (2014). Introduction to Systems Approach. Experimental Medicine. 32: 118-122.
- Kitano, H. (2014). Systems Biology and Drug Discovery: a case study. Experimental Medicine. 32: 610-615.
- Kitano, H. (2014). Introduction to Systems Science for Biologist. QIAGEN eyes. March 2014: 8-11.
4.2 Books and Other One-Time Publications
-
Ghosh, S., et al. (2013). Toward an integrated software platform for systems pharmacology. Biopharmaceutics & Drug Disposition. 34: 508–526.
-
Kitano, H. (2014). The Grand Challenge of Systems Biomedicine. World Health Summit Yearbook 2013: 38-41.
-
Abe, T. and M. Okita (2013). Flint 1.0beta4. https://flintk3.unit.oist.jp/, online. Abe, T., et al. (2014).
-
Flint 1.0 RC1. http://physiodesigner.org, online.
-
Asai, Y., et al. (2014). PhysioDesigner 1.0 RC1. http://physiodesigner.org, online.
-
Asai, Y. and L. Li (2014). PhysioVisualizer 1.0RC1. http://physiome.jp, online.
-
Asai, Y., et al. (2013). PhysioDesigner 1.0beta4. http://physiodesigner.org/, OIST, Osaka University et al.
-
Asai, Y., et al. (2013). PH Database alpha version. http://physiome.jp/phdb/index.html,online.
4.3 Oral and Poster Presentations
- Kitano, H. (2013). Systems drug discovery and Neuro-degenerative diseases. CHDI's 8th Annual Huntington's Disease therapeutics Conference: A Forum for Drug Discovery & Development. Molino Stucky Hilton Hotel. (invited)
- Kitano, H. (2013). Systems biology and systems biomedicine: integrative systems sciences and biomedical sciences. 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society. Osaka International Convention Center.(invited}
- Kitano, H. (2013). HD-Physiology Project -A Japanese Flagship Physiological Modeling Project. ICSU bio-unions satellite symposium: Multi-Scale Systems Biology. Chicheley Hall, Chicheley, UK. (invited)
- Kitano, H. (2013). Garuda Platform: An integrated inter-operability for biomedical software and data resources. COMBINE 2013. Institut Curie, Paris, France (invited)
- Kitano, H. (2014). Systems Drug Design and Software Platform for Adverse Drug Effects: Application to TKI Cardiotoxicity. American Society for Clinical Pharmacology and Therapeutics (ASCPT) 2014 Annual Meeting. Atlanta Marriott Marquis, Atlanta, Georgia, USA (invited)
- Kitano, H. (2013). System Toxicology Platform. 40th The Japanese Society of Toxicology Annaul Meeting - Symposium 7 "Toxicomics". Makuhari Messe, Chiba, Japan. (invited)
- Kitano, H. (2013). Critical View of Op-Eds on Risk of Low-Level Radiation. Exposure and Difficulties on Risk Evaluation. 40th The Japanese Scociety of Toxicology Symposium 10 - "Radiation Toxicology: Its Past, Present and Future". Makuhari Messe, Chiba, Japan. (invited)
- Kitano, H. (2013). Act Beyond Borders. KAKENHI Innovative Areas "HD-Physiology Project" young researchers' workshop. Loisir Hotel Toyohashi, Osaka. (invited)
- Asai, Y. (2013). Physiodesigner and Flint: Modeling and Simulation Tools for Multilevel Physiological Systems. The 2nd HD-Physiology International Symposium 2013, Marunouchi Plaza Hall, Tokyo.
- Asai, Y., et al. (2013). A versatile platform for multilevel modeling of physiological systems: Template/instance IEEE EMBC2013, Osaka International Convention Center.
- Kitano, H. (2013). HD-Physiology Project -A Japanese Flagship Physiological Modeling Project. The 2nd HD Physiology International Symposium: Multi-Level Systems Biology, Marunouchi MY PLAZA HALL.
- Kitano, H. (2014). What’s Open Energy Systems? . The First International Workshop on Open Energy Systems, OIST, Okinawa JAPAN.
- Oka, H., et al. (2013). Image based realistic EEG/MEG simulation in PhysioDesigner. Neuro2013, Kyoto International Conference Center, Japan.
- Asai, Y. (2014). Current status of PhysioDesigner and its future/PhysioDesigner The 8th General meeting of Integrative Multi-level Systems Biology Kyoto University.
- Asai, Y., et al. (2014). A Garuda-based cooperative system that utilizes modeling and simulation tools with the Dynamic Brain Platform. Neuro Computing Meeting (Technical Report of IEICE NC2013-86). Kyutech, Japan. 113: 111-116.
- Kitano, H. (2014). Current Status and Perspectives on Systems Drug Discovery. The 75th Coronary Circulation Symposium. Tokyo International Forum/.
- Abe, T. and Y. Asai (2013). PhysioDesigner and Flint with Garuda. Garuda workshop in ICSB2013. Copenhagen, Denmark.
- Kitano, H. (2013). Systems Biomedicine for Drug Discovery and Personalized Healthcare. Institute for Infocomm Research(I2R), Institute for Infocomm Research(I2R).
- Kitano, H. (2013). Systems Biology Platform for Drug Discovery. Talk at Imperial College, London, UK.
- Kitano, H. (2013). Systems Drug Discovery and Integrated Software Platform. ETH Zurich IMSB, Zurich, Switzerland.
- Kitano, H. (2013). Systems Biology and Software Platform. Nature Publishing Group Asian Academic Publishing team meeting, Sheraton Miyako Hotel.
- Asai, Y. (2013). Special Lecture for Niigata University Global Human Resource Development “Internationality from Okinawa” Niigata University.
- Asai, Y. and H. Kitano (2013). Getting started in Physiological Modeling with PhysioDesigner. Mitsubishi Tanabe Pharma
- Asai, Y. and H. Kitano (2013). Getting started in Physiological Modeling with PhysioDesigner. Kyowa Kirin.
- Asai, Y. and H. Kitano (2014). Physiological Modeling of Kidney. Kyowa Kirin.
- Kitano, H. (2013). Biomarker evaluation using systems biology approach. Nikkei Biotech/Nikkei Biotech ONLINE Professional Seminar “The advent of clinical biomarker age to learn from examples” KOKUYO Hall.
- Kitano, H. (2013). Biomarker evaluation using systems biology approach. Lecture hosted by Tohoku Medical Megabank Organization, Gonryo Kaikan Hall.
- Kitano, H. (2013). Multi-layer hierarchical physiology: from model-based drug discovery to clinical decision support. The 15th KMU Research and Development Seminar Kanazawa Medical University Hospital, Kanazawa.
- Kitano, H. (2013). Current Status and Perspectives on Systems Drug Discovery. Asubio Phrma Interoffice Seminar Asubio Pharma Co, Ltd.
- Asai, Y., et al. (2013). Multilevel modeling using SBML and PHML on PhysioDesigner. ICSB2013. Copenhagen, Denmark.
-
Asai, Y., et al. (2013). A Versatile Platform for Multilevel Modeling of Physiological Systems: SBML-PHML Hybrid Modeling and Simulation. . JBMES2013. Kyutech, Japan.
-
Asai, Y., et al. (2013). PhysioDesigner and Flint: a platform for multilevel modeling of physiological systems and simulation. ICCB2013. Leuven, Belgium. .
-
Asai, Y., et al. (2013). Flint K3: Cloud-based simulation service powered by Flint. The 2nd HD-Physiology International Symposium 2013. Marunouchi Plaza Hall, Tokyo.
-
Asai, Y., et al. (2013). PhysioDesigner for Multilevel Neural System Modeling. Neuroinformatics2013. Karolinska Institutet, Sweden.
5. Intellectual Property Rights and Other Specific Achievements
- Kitano, H., et al. (2013). Network model integration device, network model integration system, network model integration method, and computer program product. J. P. Office. Japan, OIST. SBI.
6. Meetings and Events
6.1 The First International Symposium for Open Energy Systems
- Date: January 14-15, 2014
- Venue: OIST Campus (Seminar Room B250, center Bldg.)
- Speakers:
- Prof. Rajendra Kumar Pachauri, The Intergovernmental Panel on Climate Change (IPCC)
- Prof. Dave Griggs, ClimateWorks Australia/Monash Sustainability Institute
- Dr. Stephen Ronan, ALSTOM
- Mr. Dennis Teranishi, Pacific International Center for High Technology Research, Hawaii
- Dr. Thomas Rivera, Orange Labs Networks
- Mr. Yuki Shimazu, METI
- Mr. Toshiyuki Hayashi, JICA
- Prof. Toru Morotomi, Kyoto University
- Prof. Kenji Tanaka, The University of Tokyo
- Dr. Hiroshi Fujiwara, Broadband Tower Inc.
- Dr. Mario Tokoro, Sony CSL
- Prof. Hiroaki Kitano, OIST/SonyCSL
- Co-organizer: Sony Computer Science Laboratories, Inc. (Sony CSL)
- Supported by:
- Australian Embassy, Tokyo
- Embassy of France in Japan
- Victoria Government Business Office in Tokyo
- Japan International Cooperation Agency (JICA)
- Okinawa Prefecture
6.2 Seminar
-
Date: October 29, 2013
-
Venue: OIST Campus (B403, Lab1)
-
Speaker: Professor Malcolm Walkinshaw, Chair of Structural Biochemistry/Institute of Structural and Molecular, Biology/School of Biological Sciences/University of Edinburgh
6.3 Seminar
- Date: Ammrch 18, 2014
- Venue: OIST Campus (C015, Lab1)
- Speakers:
- Professor Koji Koyamada, Institute for Liberal Arts and Sciences, Kyoto University
- Dr. Naohisa Sakamoto, Institute for Liberal Arts and Sciences, Kyoto University



