FY2019 Annual Report
Nucleic Acid Chemistry and Engineering Unit
Associate Professor Yohei Yokobayashi

Abstract
FY2019 marks the fifth year since the unit was established at OIST. Our efforts to engineer functional nucleic acids encompass systems that operate in test tubes, mammalian cells, and artificial cells. A number of key projects initiated at OIST came to fruition as publications some of which are summarized below. Furthermore, multiple unit members successfully competed for KAKENHI grants.
1. Staff
- Dr. Keisuke Fukunaga, Researcher
- Dr. Samuel Hauf, Researcher
- Dr. Narae Kim, Researcher
- Dr. Yoko Nomura, Science and Technology Associate
- Dr. Vyankat Sontakke, Researcher
- Dr. Kei Takahashi, Researcher
- Kamila Mustafina, Graduate Student
- Takeshi Tabuchi, Graduate Student
- Rachapun (Gear) Rotrattanadumrong, Graduate Student
- Shivang Joshi, Research Intern
- Mayumi Suzuki, Technical Staff
- Hitomi Shinzato, Research Unit Administrator
- Takako Kai, Research Assistant
- Yayoi Maehara, Research Assistant
(As of 3/31/2020)
2. Collaborations
2.1 Applications of cell-free riboswitches
- Description: Synthetic riboswitch applications in cell-free systems.
- Type of collaboration: Joint research
- Researchers:
- Professor Tomoaki Matsuura, Osaka University
2.2 Applications of aptamers for biosensors
- Description: Using DNA aptamers to sense proteins using bionanosensors
- Type of collaboration: Joint research
- Researchers:
- Professor Mukhles Sowwan, OIST (Nanoparticles by Design Unit)
2.3 Collaboration with an industrial partner
- Undisclosed collaboration with an industrial partner.
3. Activities and Findings
3.1 Riboswitches in Cell-Free Systems and Artificial Cells
Artificial cell models composed of lipid vesicles (liposomes) encapsulating an in vitro (cell-free) transcription-translation system (IVTT) are emerging as a part of the efforts to synthesize cells from bottom up. Researchers have constructed artificial cells that mimic various biological properties such as DNA replication, cell-cell communication, and cell division. While the ultimate goal is to synthesize artificial cells (from scratch) that replicate and evolve like natural cells, this field is still at an early stage where artificial cells are being equipped with various biological and genetic machineries individually. We recognized that artificial cells are still limited in their ability to genetically respond to chemical signals in and out of cells. Such chemical interfaces are critical for artificial cells to sense and respond to a dynamic environment by regulating gene expression. Consequently, we proceeded to demonstrate the utility of riboswitches as a chemical interface in artificial cells by developing an RNA aptamer for a novel (desired) signal, engineering cell-free riboswitches, and using them in artificial cells to confer new phenotypes that are triggered by the aptamer ligand (Fig. 1).
Histamine was chosen as the chemical input. We viewed histamine as a model chemical (inflammation) signal that could be sensed by artificial cells, which in turn respond to histamine by expressing an arbitrary protein that confer a specific phenotype.
First, we successfully selected an RNA aptamer for histamine with a Kd of 370 nM. Second, the aptamer was used to engineer histamine-activated riboswitches with ON/OFF ratios as high as 30 (Fig. 1B). Finally, in collaboration with Prof. Tomoaki Matsuura at Osaka University, we constructed artificial cells containing riboswitch-controlled genes (Fig. 1C). By controlling the expression of a nanopore-forming protein (α-hemolysin), we demonstrated the histamine-triggered release of a small molecule payload (model drug) (Fig. 1E). Additionally, histamine-triggered self-destruction was demonstrated using phospholipase C as a model of an emergency kill-switch. The modularity of riboswitches further adds to the flexibility in programming artificial cell behavior. A provisional patent application has been filed for the histamine aptamer, with possible applications for biosensors, as histamine in food samples are routinely monitored for food poisoning detection.
Figure 1: Cell-free histamine riboswitch in artificial cells. (A) Schematic illustration of the riboswitch mechanism. (B) Histamine-dependent expression of mCherry by the riboswitch H2. (C) Schematic illustration of the riboswitch-regulated artificial cells. (D)-(E) Artificial cells expressing mCherry (D) or α-hemolysin under the control of the H2 riboswitch. OA647 (45 kDa) is a fluorescently labeled ovalbumin protein used to image the artificial cell volumes. Calcein (623 Da) serves as a model small molecule payload. +Histamine: 10 mM histamine added to the outer solution. From Dwidar et al. 2019.
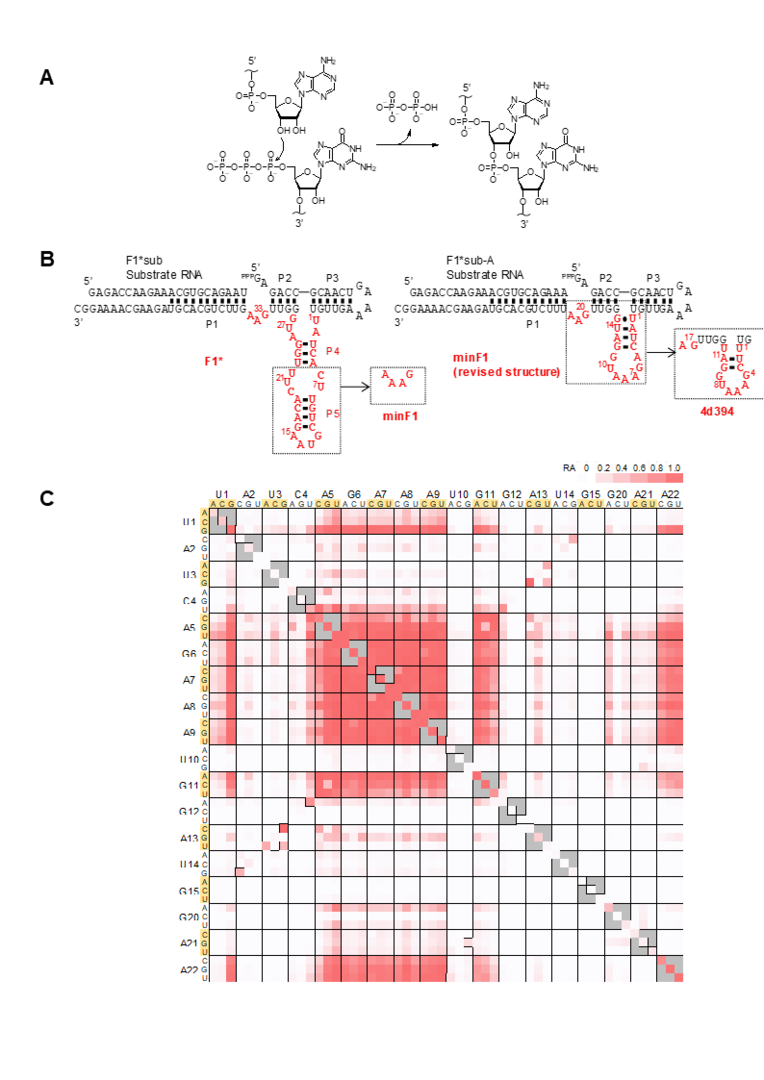
3.2 Minimization of an RNA Ligase Ribozyme
We used the deep sequencing-based assay to analyze an important class of laboratory-evolved ribozymes, an RNA ligase ribozyme that catalyzes the template-directed ligation of an RNA fragment’s 3’-OH group and the 5’-triphosphorylated RNA fragment (Fig. 2A). As the template-directed nature of the reaction allows for the propagation of genetic information, and the ligation chemistry is analogous to the modern-day RNA and DNA polymerases, these RNA ligase ribozymes may resemble what may have existed the RNA world. In fact, one such ligase F1 was engineered to undergo crosscatalytic self-replication without any protein factors (Robertson & Joyce Chem Biol 2014; 21, 238-245.).
|
Fig. 2 Deep mutational analysis and minimization of an RNA ligase ribozyme (A) Ligation chemistry. (B) Sequence and structures of F1* (parental), minF1, and 4d394 ligases. (C) Double mutation matrix of minF1. From Nomura & Yokobayashi 2019. |
We decided to perform deep mutational analysis of an F1-derived ribozyme (F1*) using deep sequencing. In addition to single and double substitutions, all possible single and double deletion mutants were synthesized and analyzed. This analysis led us to identify a nonessential region within the catalytic core, which was removed and confirmed to be catalytically active (minF1). minF1 was further analyzed by deep mutational analysis which revealed base-base interactions previously unknown and slow but still active mutants with 4-base deletions (4d394) (Fig. 2B,C). The minimized catalytic core of only 18-nt suggests that relatively small RNA sequence motifs may be able to catalyze chemical reactions, albeit slowly. However, such small but slow ribozymes may have been overlooked in the conventional laboratory selection/evolution experiments in which faster variants quickly dominate slower variants. We were able to discover such motifs thanks to the high throughput assay and systematic analysis enabled by deep sequencing. Because smaller motifs have a higher probability of emergence in the primitive RNA world in which replication efficiency and accuracy were modest, such small catalytic motifs may be worth further investigations.
3.3 Chemically-regulated RNA viral vector
While existing mammalian riboswitches leave some room for improvement in performance, it is important to start exploring the potential applications of these RNA devices. Using prototypical a guanine-responsive riboswitch (GuaM8HDV developed in our group) as a model, we have started applying riboswitches for more practical applications.
Vesicular stomatitis virus (VSV) is a unique virus with potential clinical applications. VSV is a nonsegmented, negative-sense, cytoplasmic RNA virus that replicates without DNA intermediates; therefore, there is no risk of genomic integration. Furthermore, VSV is virtually nonpathogenic to humans and can stably (>weeks) and strongly express exogenous genes. VSV vectors have been investigated for applications as oncolytic viruses and vaccine vectors.
We used a guanine-responsive riboswitch (GuaM8HDV) to reversibly regulate VSV vector replication and transgene expression over the course of 12 days (Fig. 3). We also identified the best combination of genes to be regulated by the riboswitch.
|
Fig. 3 Reversible gene regulation using riboswitch-controlled VSV vectors. (A) BHK-21 cells were infected with VSV expressing NanoLuc luciferase, and guanine was added and removed every 48 h. Riboswitch-regulated VSV (red) and unregulated VSV (black). (B) BHK-21 cells infected with riboswitch-regulated VSV expressing EGFP with and without guanine. From Takahashi & Yokobayashi 2019. |
In a separate project, we are using VSV engineered to express differentiation factors under the control of riboswitches to regulate the differentiation of embryonic stem (ES) cells. The use of non-integrating RNA virus vectors to control stem cell differentiation is highly desirable for clinical applications. We are investigating if riboswitches can improve the efficiency and precision of stem cell differentiation by modulating the expression levels and/or timing of differentiation factors.
3.4 Self-Powered RNA Nanomachine Driven by Metastable Structure
Most current synthetic RNA devices such as those described above are designed to function in their thermodynamically stable structures. Aptamer-ligand interaction, for example, provides the driving force to alter the thermodynamically stable structure, resulting in functions such as self-cleavage and gene regulation. In reality, however, some RNAs misfold during transcription as RNAs are folded cotranscriptionally. Some misfolded structures are kinetically (but not thermodynamically) stable, which may explain some of the unreported failures of RNA design efforts.
We were inspired by the fact that some cells actively exploit such cotranscriptionally folded and kinetically stable (metastable) structures. Consequently, we set out to design de novo RNA devices that adopt metastable structures cotranscriptionally. Furthermore, we aimed to control the device’s relaxation to the thermodynamically stable structure.
We first designed robust cotranscriptionally-generated metastable RNA structures that persist for hours at ambient temperature. These structures almost entirely relaxed to the thermodynamically stable structures after heat denaturation. The devices were designed so that the relaxed (thermodynamically stable) structures become fluorescent. We then modified the devices so that the relaxation was catalyzed by specific RNA sequences. Transient hybridization of the trigger RNA exposed an internal toehold sequence through which structural rearrangement was initiated isothermally. The trigger RNA was released upon relaxation, allowing it to catalyze additional structural rearrangements. This metastable RNA device thus functions as an RNA sensor with a signal amplifier and an optical readout (Fig. 4). More importantly, the device is self-powered by the energy stored in its metastable structure.
|
Fig. 4 Self-powered RNA nanomachine driven by a metastable structure. (A) Schematic illustration of the RNA nanomachine wMT8. Upon in vitro transcription from a DNA template, wMT8 is almost exclusively folded (cotranscriptionally) into the branched metastable structure. This structure is stable for hours at room temperature and >17 h at 4 °C. The RNA relaxes to the thermodynamically stable rod-like structure which also includes the functional Broccoli aptamer that becomes fluorescent upon binding DFHBI-1T. The relaxation can be triggered by heat denaturation-renaturation, or catalytically via a short trigger RNA that transiently hybridizes with wMT8. (B) Catalytic relaxation of wMT8 (1000 nM) in the presence of the trigger RNA. NC: no catalyst. From Kobori et al. 2019. |
3.5 Novel Mammalian Riboswitch Architecture: Tandem Aptamer-Ribozyme Fusion
Aptazymes have been used extensively to enable small molecule-responsive gene regulation in mammalian cells. However, the general design strategy of aptazymes has mostly been limited to inserting an aptamer to an existing stem loop structure of a self-cleaving ribozyme. This aptamer-in-ribozyme architecture often necesitates extensive and labor intensive screening of variants in the "communication module", or the sequence that interface the aptamer and the host ribozyme.
We devised a new aptazyme architecture that place an aptamer and a ribozyme in tandem, with a rationally-tunable anti-ribozyme sequence (Fig. 5A). In this configuration, the ligand-bound aptamer and the ribozyme structures are mutually exclusive. In the absence of the ligand, the ribozyme structure is more stable, which results in mRNA cleavage and low protein expression (OFF). In the presence of the ligand, the aptamer-ligand structure forms resulting in the inhibition of ribozyme cleavage (ON). We envisioned that we could optimize the riboswitch performance by semi-rational tuning of the anti-ribozyme sequence.
We demonstrated the design strategy by using a guanine aptamer fused to a twister ribozyme. After several rounds of optimization, we obtained several riboswitches with an ON/OFF ratio > 6.0 (Fig. 5)
|
Fig. 5 Optimization of P3apt stability in the aptazyme architecture by introducing mismatches. (A) Representative Apt and Rz structures illustrated for the 7c variant. (B) Normalized EGFP expression levels of the riboswitch variants in the absence and presence (250 mM) of guanine. EGFP: empty vector with no aptazyme. (C) Dose-dependent EGFP expression of 7c4x. The data are fitted to the Hill equation which is shown as the solid curve. (D) Fluorescence micrographs of the cells shown in (C). (B)-(C): The data are averages of 3 replicate samples and the error bars represent S.D. From Mustafina et al. 2020. |
4. Publications
4.1 Journals
- Mustafina K, Fukunaga K, Yokobayashi Y. Design of Mammalian ON-Riboswitches Based on Tandemly Fused Aptamer and Ribozyme. ACS Synth Biol 2020; 9: 19-25.
- Dwidar M, Yokobayashi Y. Development of a histamine aptasensor for food safety monitoring. Sci Rep 2019; 9: 16659.
- Danielson E, Dhamodharan V, Porkovich A, Kumar P, Jian N, Ziadi Z, Grammatikopoulos P, Sontakke VA, Yokobayashi Y, Sowwan M. Gas-Phase Synthesis for Label-Free Biosensors: Zinc-Oxide Nanowires Functionalized with Gold Nanoparticles. Sci Rep 2019; 9: 17370.
- Nomura Y, Yokobayashi Y. Systematic minimization of RNA ligase ribozyme through large-scale design-synthesis-sequence cycles. Nucleic Acids Res 2019; 47: 8950-8960.
- Takahashi K, Yokobayashi Y. Reversible Gene Regulation in Mammalian Cells Using Riboswitch-Engineered Vesicular Stomatitis Virus Vector. ACS Synth Biol 2019; 8: 1976-1982.
- Saito H, Yokobayashi Y. Editorial overview: Mammalian synthetic biology: from devices to multicellular systems. Curr Opin Chem Biol 2019; 52: 1-2.
- Yokobayashi Y. Aptamer-based and aptazyme-based riboswitches in mammalian cells. Curr Opin Chem Biol 2019; 52: 72-78.
- Dwidar M, Seike Y, Kobori S, Whitaker C, Matsuura T, Yokobayashi Y. Programmable Artificial Cells Using Histamine-Responsive Synthetic Riboswitch. J Am Chem Soc 2019; 141: 11103-11114.
- Kobori S, Nomura Y, Yokobayashi Y. Self-powered RNA nanomachine driven by metastable structure. Nucleic Acids Res 2019; 47: 6007-6014.
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- Yokobayashi Y. Designing RNA Devices in Test Tubes and Cells, 46th International Symposium on Nucleic Acids Chemistry, Tokyo, Japan, Oct (2019). Invited talk.
- Yokobayashi Y. 人工細胞でも使える(?)RNAデバイスの作成 (RNA Devices for Potential Applications in Artificial Cells), 「細胞を創る」研究会 12.0 (Saibou wo Tsukuru Kenkyu Kai 12.0 /Annual Meeting of Japanese Society of Cell Synthesis Research), Ehime, Japan Oct (2019). Invited talk.
- Yokobayashi Y. Department of Applied Chemistry and Biotechnology, Chiba University, Chiba, Japan, Sep (2019). Invited talk.
- Yokobayashi Y. Synthetic Mammalian Riboswitches, OIST-NTHU Joint Seminar 2019, Okinawa, Japan, Aug (2019). Invited talk.
- Yokobayashi Y. Engineering RNA Gene Switches, AS x OIST Joint Symposium 2019, Okinawa, Japan, Apr (2019). Invited talk.
- Yokobayashi Y, Dwidar M, Seike Y, Kobori S, Whitaker C, Matsuura T. Expanding Chemical Interface of Artificial Cells by Synthetic Riboswitch, Cell Free Systems Conference, Boston, U.S.A., Dec 4-6 (2019). Poster.
- Tabuchi T, Yokobayashi Y. High-Throughput Screening of Cell-Free Riboswitches by Droplet Sorting, Cell Free Systems Conference, Boston, U.S.A., Dec 4-6 (2019). Poster.
- Nomura Y, Yokobayashi Y. Systematic Minimization of RNA Ligase Ribozyme Through Large-Scale Design-Synthesis-Sequence Cycles, The 46th International Symposium on Nucleic Acids Chemistry, Tokyo, Japan, Oct 29-31 (2019). Poster.
- Takahashi K, Yokobayashi Y. Reversible Gene Regulation Using Riboswitch-Engineered Vesicular Stomatitis Virus Vector, The 46th International Symposium on Nucleic Acids Chemistry, Tokyo, Japan, Oct 29-31 (2019). Poster.
- Mustafina K, Fukunaga, K, Yokobayashi Y. Design of Mammalian ON-riboswitches Based on Tandemly Fused Aptamer and Ribozyme, The 46th International Symposium on Nucleic Acids Chemistry, Tokyo, Japan, Oct 29-31 (2019). Poster.
- Takahashi K, Yokobayashi Y. Reversible Gene Regulation in Mammalian Cells Using Riboswitch-Engineered Vesicular Stomatitis Virus Vector, Sixth International Mammalian Synthetic Biology Workshop (mSBW 6.0), Evanston, IL, U.S.A., May 17-19 (2019). Poster.
5. Intellectual Property Rights and Other Specific Achievements
A provisional patent application for the development and applications of a histamine aptamer was filed.
6. Meetings and Events
6.1 E-field Modulation of Molecular Assemblies and Functions by Engineering Electrostatic Interactions
- Date: November 5, 2019
- Venue: OIST Campus Lab1
- Speaker: Prof. Yoshimitsu Itoh (The University of Tokyo)
7. Other
Research intern: Shivang Joshi (Imperial College London) January 2020-
Rotation student: Yatzu Chiu (September-December 2019)
External funding awarded (*continuing)
- PI: Yohei Yokobayashi, JSPS KAKENHI Kiban B (19H02855), 2019-2022
- PI: Yohei Yokobayashi, JSPS KAKENHI Challenging Research (Exploratory) (18K19944), 2018-2021*
- PI: Kei Takahashi, JSPS KAKENHI Early-Career Scientists (19K15988), 2019-2021
- PI: Keisuke Fukunaga, JSPS KAKENHI Early-Career Scientists (19K15701), 2019-2021
- PI: Mohammed Dwidar, JSPS KAKENHI Early-Career Scientists (19K16093), 2019-2022
- PI: Yoko Nomura, JSPS KAKENHI Kiban C (19K02308), 2019-2022
Services
Local Committee Member, The 20th International Conference on Systems Biology, Nov 2019, Okinawa, Japan