FY2017 Annual Report
Nucleic Acid Chemistry and Engineering Unit
Associate Professor Yohei Yokobayashi
Abstract
In FY2017, we applied the deep sequencing based high-throughput assay of ribozymes to construct various ribozymes and riboswitches for controlling gene expression. The methodology was also applied to a DNA enzyme or deoxyribozyme. Other progress include engineering a more stable and active firefly luciferase in collaboration with UC Davis and gene regulation in a predatory bacterium using riboswitches.
1. Staff
- Dr. Mohammed Dwidar, Researcher
- Dr. Narae Kim, Researcher
- Dr. Yoko Nomura, Science and Technology Associate
- Dr. Kei Takahashi, Researcher
- Dr. Dhamodharan Venugopal, Researcher
- Kamila Mustafina, Graduate Student
- Dalmira Merzhakupova, Graduate Student
- Takeshi Tabuchi, Graduate Student
- Bidesh Chatterjee, Rotation Student
- Laura Croenen, Technical Staff
- Hitomi Shinzato, Research Unit Administrator
(As of 3/31/2018)
2. Collaborations
2.1 In vivo 3D Imaging Using Bioluminescent Gene Reporters and MRI
- Description: We have been collaborating with two groups at University of California, Davis to develop a novel MRI probes to monitor gene expression in living animals.
- Type of collaboration: Joint research
- Researchers:
- Professor Angelique Louie, University of California, Davis
- Professor Jared Shaw, University of California, Davis
2.2 Collaboration with an industrial partner
- Undisclosed collaboration with an industrial partner.
3. Activities and Findings
3.1 Applications of Deep Sequencing to Analyze and Engineer Nucleic Acid Enzymes
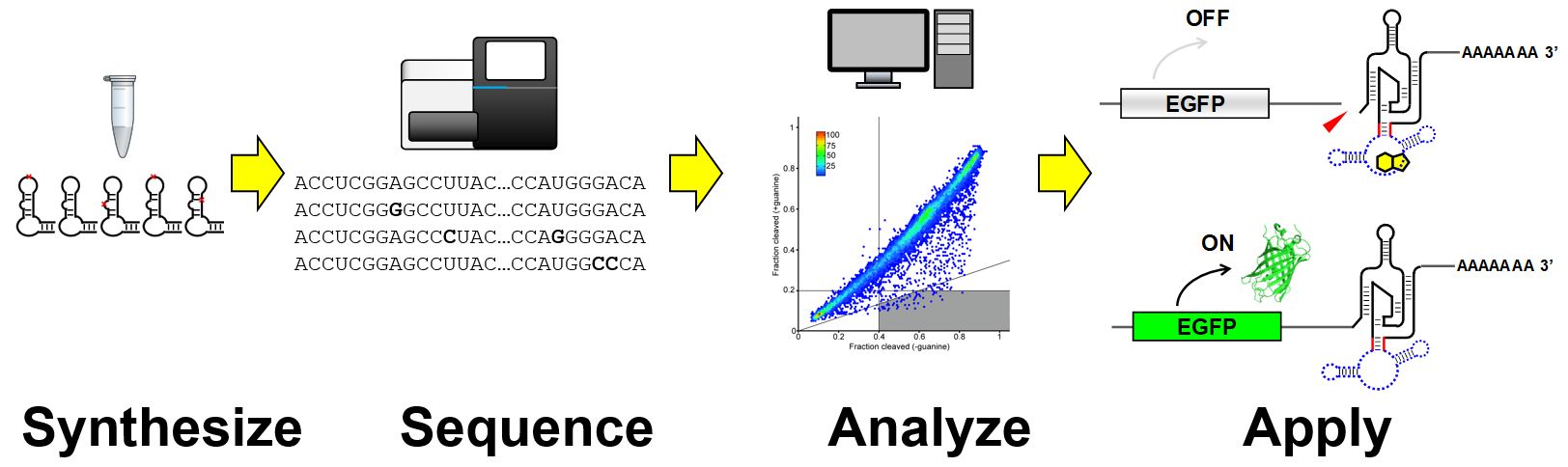
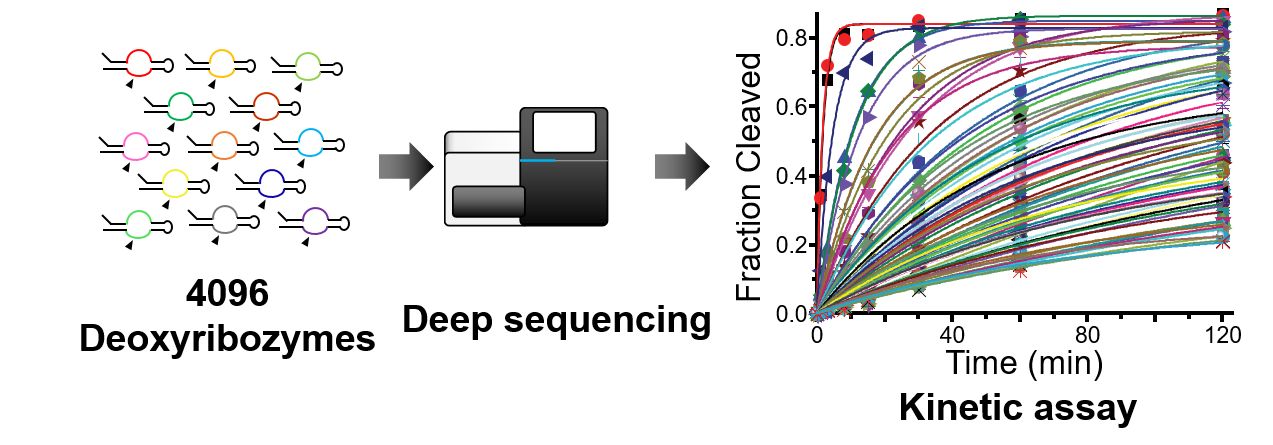
Building on the foundational methodologies developed during FY2015-16, we demonstrated various applications of deep sequencing to not just analyze but also to engineer nucleic acid enzymes (i.e. ribozymes and deoxyribozymes). Our approach harnesses the power of high-throughput sequencing to quantitatively assay ribozyme and deoxyribozyme activities in a massively parallel fashion. We used the method to analyze a large number of locally mutated ribozymes to probe the sequence-function relationship as well as to generate a simple RNA platform to fine-tune gene expression in mammalian cells (Kobori & Yokobayashi 2018). We also developed a novel aptazyme architecture that connects an aptamer and a ribozyme in tandem (Kobori et al. 2017). Furthermore, we extended the deep sequencing assay to directly measure the activities of ribozyme populations in mammalian cells (Nomura et al. 2017), allowing direct assay of ribozyme activity in living cells rather than in test tubes. Finally we applied the method for the first time to a DNA enzyme (deoxyribozyme) and performed a detailed kinetic analysis for a large set of mutants to gain fundamental insights (Dhamodharan et al. 2017). This series of work demonstrates the robustness and practical utility of our sequencing-based assays of nucleic acid enzymes, and sets the stage for the future efforts to engineer more complex and useful DNA and RNA devices.
3.2 Synthetic Bacterial Riboswitches
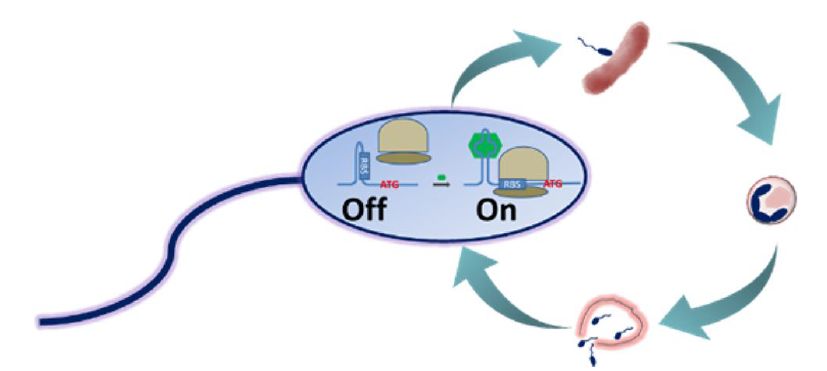
Bdellovibrio bacteriovorus by a synthetic riboswitch.
Riboswitches are chemically responsive gene regulatory elements that use RNA for both molecular recogniation (sensing) and gene regulation. Synthetic riboswitches have promising applications in biotechnology and medicine by enabling cells to respond to arbitrary chemical cues to control any genes of interest. In FY2017, we continued our efforts to develop riboswitches that respond to a small molecule inflammatory signal for potential probiotics applications. Specifically, we biochemically characterized a novel RNA aptamer and used it to engineer synthetic riboswitches using in vitro transcription/translation system. Data have been collected for future patent application for the aptamer.
We also developed riboswitches that function in the predatory bacterium Bdellovibrio bacteriovorus which is known to predate a wide spectrum of gram-negative bacteria, including pathogenic strains. We successfully demonstrated that we can activate gene expression by a small molecule (theophylline) via previously reported riboswitches. The riboswitch was further inserted into the bacterial genome to control a gene that is involved in predation. The predation behavior of the recombinant bacteria was thus controlled by the chemical signal (Dwidar & Yokobayashi 2017).
mutant with enhanced activity
and stability
3.3 Protein Engineering of Firefly Luciferase
In collaboration with Prof. Angelique Louie and Prof. Jared Shaw, we have been attempting to activate optically-responsive magnetic resonance imaging (MRI) probles using light emitted from a genetically encoded luciferase. Such a system can be used, for example, to track cancer cells in vivo using MRI which offers superior deep tissue imaging and resolution compared to optical imaging or CAT. Our task was to engineer luciferases that are both more active and stable than the wild-type luciferase. The firefly (Photinus pyralis) luciferase is widely used for bioluminescence imaging but it is rather unstable, and is deactivated in minutes at 37 °C in vitro. Also, its intracellular activity is limited by the low concentration of its substrate (luciferin). Therefore, we combined mutations previously identified to enhance stability and sensitivity (Km) to luciferin, and discovered that the new mutant luciferase retains both favorable properties in vitro (Pozzo et al. 2018). Efforts are underway to use this luciferase mutant to activate optically responsive MRI probes.
4. Publications
4.1 Journals
- Kobori S, Takahashi K, Yokobayashi Y. Deep Sequencing Analysis of Aptazyme Variants Based on a Pistol Ribozyme. ACS Synth Biol 2017; 6: 1283–1288.
- Dwidar M, Yokobayashi Y. Controlling Bdellovibrio bacteriovorus Gene Expression and Predation Using Synthetic Riboswitches. ACS Synth Biol 2017; 6: 2035-2041.
- Dhamodharan V, Kobori S, Yokobayashi Y. Large Scale Mutational and Kinetic Analysis of a Self-Hydrolyzing Deoxyribozyme. ACS Chem Biol 2017; 12: 2940-2945.
- Nomura Y, Chien H-C, Yokobayashi Y. Direct screening for ribozyme activity in mammalian cells. Chem Commun 2017; 53: 12540-12543.
- Kobori S, Yokobayashi Y. Analyzing and Tuning Ribozyme Activity by Deep Sequencing to Modulate Gene Expression Level in Mammalian Cells. ACS Synth Biol 2018; 7: 371-376.
- Pozzo T, Akter F, Nomura Y, Louie AY, Yokobayashi Y. A Firefly Luciferase Mutant with Enhanced Activity and Thermostability. ACS Omega 2018; 3: 2628–2633.
4.2 Books and other one-time publications
- Yokobayashi Y. Small Molecule‐Responsive RNA Switches (Bacteria): Important Element of Programming Gene Expression in Response to Environmental Signals in Bacteria. in Synthetic Biology: Parts, Devices and Applications pp181-188, eds: Smolke C, Lee SY, Nielsen J, Stephanopoulos G, 2018; Wiley-VCH (book chapter)
4.3 Oral and Poster Presentations
- Kobori S, Yokobayashi Y. High-throughput mutational analysis and engineering of self-cleaving ribozymes by sequencing, 253rd American Chemical Society National Meeting, San Francisco, USA, Apr 2-6 (2017). Poster
- Venugopal D, Kobori S, Yokobayashi Y. Mutational studies of a deoxyribozyme by high-throughput sequencing, 253rd American Chemical Society National Meeting, San Francisco, USA, Apr 2-6 (2017). Poster
- Kobori S, Nomura Y, Takahashi K, Yokobayashi Y. Engineering and comprehensive analysis of RNA switch by next generation sequencing, 5th NGS Field Meeting, Sendai, Japan, May 22-24 (2017). Poster
- Dwidar M, Yokobayashi Y. Controlling Bdellovibrio bacteriovorus gene expression and predation using synthetic riboswitches, 2017 Synthetic Biology: Engineering, Evolution & Design (SEED), Vancouver, Canada, Jun 20-23 (2017). Poster
- Yokobayashi Y. Large Scale Mutational Analysis of Nucleic Acid Enzymes by Deep Sequencing, 7th Neobio Molecule Symposium, Naha, Japan, Jul 6 (2017). Invited talk
- Venugopal D, Kobori S, Yokobayashi Y. Mutational and Kinetic Profiling of a Deoxyribozyme by High-Throughput Sequencing, The 44th International Symposium on Nucleic Acids Chemistry, Tokyo, Japan, Nov 14-16 (2017). Poster
- Nomura Y, Chien H-C, Yokobayashi Y. Direct Screening of Self-Cleaving Ribozyme Activity in Mammalian Cells, The 44th International Symposium on Nucleic Acids Chemistry, Tokyo, Japan, Nov 14-16 (2017). Poster
- Yokobayashi Y. Engineering RNA Devices to Control Gene Expression, Department of Chemistry and Biotechnology, The University of Tokyo, Tokyo, Japan, Dec 5 (2017). Invited talk
- Dwidar M, Yokobayashi Y. Controlling gene expression in a predatory bacterium using synthetic riboswitches, ICBE Asia 2018, Singapore, Jan 8-10 (2018). Oral presentation
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 Molecular Technologies to Analyze and Control Dynamic Cellular Functions
- Date: October 5, 2017
- Venue: OIST Campus Lab1
- Speaker: Dr. Shinsuke Sando (The University of Tokyo)
7. Other
Hosted a doctoral student intern from Osaka University (Mr. Keisuke Morita) from Jan-Mar, 2018.
Taught an intensive graduate course at The University of Tokyo in December 2017.



