FY2018 Annual Report
Nucleic Acid Chemistry and Engineering Unit
Associate Professor Yohei Yokobayashi

Abstract
In FY2018, we continued to develop and analyze various engineered RNA devices to controll gene expression in living cells. Specifically, we synthesized a photocaged derivative of guanine which was used to optically control gene expression in Escherichia coli and mammalian cells via riboswitches. We also developed a signal amplification system that achieves extremely high dynamic ranges by riboswitch-controlled gene expression. Furthermore, we started applying our synthetic RNA devices to cell-free systems through a new collaboration with Osaka University and a newly-funded KAKENHI project. Although undisclosable at this point, a collaboration with an industrial partner has also advanced to a critical stage.
1. Staff
- Dr. Mohammed Dwidar, Researcher
- Dr. Keisuke Fukunaga, Researcher
- Dr. Narae Kim, Researcher
- Dr. Yoko Nomura, Science and Technology Associate
- Dr. Kei Takahashi, Researcher
- Dr. Dhamodharan Venugopal, Researcher
- Kamila Mustafina, Graduate Student
- Takeshi Tabuchi, Graduate Student
- Rachapun (Gear) Rotrattanadumrong, Graduate Student
- Laura Croenen, Technical Staff
- Hitomi Shinzato, Research Unit Administrator
- Takako Kai, Research Assistant
(As of 3/31/2019)
2. Collaborations
2.1 Applications of synthetic riboswitches
- Description: Exploration of synthetic riboswitch applications in cell-free systems.
- Type of collaboration: Joint research
- Researchers:
- Professor Tomoaki Matsuura, Osaka University
2.2 Collaboration with an industrial partner
- Undisclosed collaboration with an industrial partner.
3. Activities and Findings
3.1 Optical Control of Gene Expression in Living Cells
(pc-G) compound (Dhamodharan et al. 2018).
Optogenetics has greatly accelerated several areas of biological research by allowing regulation of biological molecules such as ion channels and transcription factors by light. We developed a novel system to control gene expression in bacteria and mammalian cells without an engineered protein. Specifically, we designed and synthesized a photocaged guanine derivative (Figure 1) by masking the O6 of guanine with a photolabile group. This photocaged guanine (pc-G) was deprotected by UV irradiation to produce free guanine which can regulate the guanine-responsive riboswitch in the targeted cells. We demonstrated UV-triggered repression of GFP expression in HEK 293 cells (a human cell line) (Figure 2) and in E. coli.
Figure 2: UV irradiation deprotects pc-G and generates free guanine which binds to the guanine aptamer in the 3' UTR of the EGFP mRNA. The associated ribozyme is activated and self-cleaves upon guanine binding, resulting in mRNA degradation and repression of EGFP protein expression (left). HEK 293 cells transfected with EGFP-riboswitch and mCherry expression vectors are treated with pc-G. UV irradiation results in suppression of EGFP expression (right). From Dhamodharan et. al. 2018
3.2 Signal Amplification of Riboswitch Output
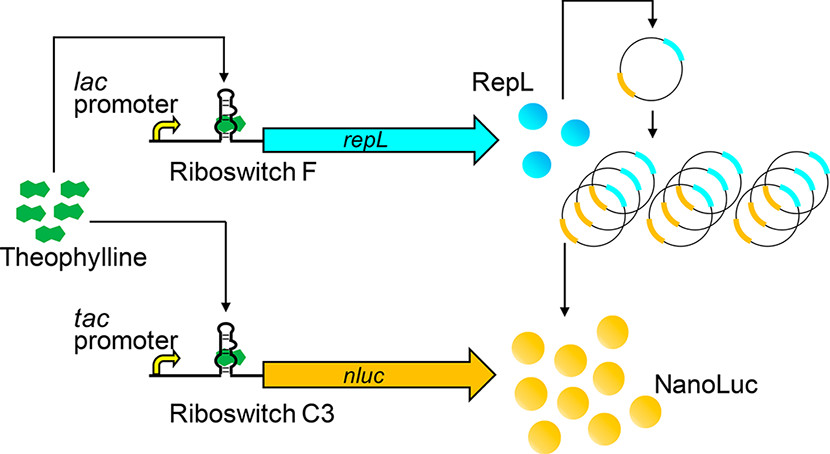
Riboswitches can control gene expression in bacteria (and in other cells) in response to specific chemical inputs through small molecule-RNA interaction. The lack of protein factors involved and the potential to engineer RNA aptamers for diverse ligands make riboswitches promising tools to control gene expression for synthetic biology applications. However, one of the major challenges of synthetic riboswitches is their relatively low dynamic range (ON/OFF ratio). Although ON/OFF ratios of ~100 have been reported in few cases, typical riboswitches exhibit ON/OFF ratios of less than 10, making them less appealing for demanding applications. We achived ON/OFF ratios as high as 3900 by using riboswitches to control plasmid replication as well as transgene expression (Figure 3). Although the riboswitches themselves exhibit moderate ON/OFF ratios of 5 to 13, simultaneous regulation of both plasmid replication and transgene expression results in synergistic enhancement of dynamic range.
3.3 Development of Histamine-Responsive Riboswitch in Artificial Cells
In collaboration with Prof. Matsuura at Osaka University, we started to construct artificial cells that respond to histamine by activating expression of a protein within liposomes that encapsulate reconstituted in vitro transcription-translation (IVTT) system (PURE System). During FY18, we isolated and characterized an RNA aptamer that selectively binds histamine, an inflammatory marker and a neurotransmitter. We further designed riboswitches based on the aptamer. The Matsuura group has been using the riboswitch we developed to contruct artificial cells that respond to histamine by activating expression of arbitrary proteins. These artificial cells serve as models for chemically responsive protocells as well as potential applications in drug delivery vehicles.
4. Publications
4.1 Journals
- Dhamodharan V, Nomura Y, Dwidar M, Yokobayashi Y. Optochemical control of gene expression by photocaged guanine and riboswitches. Chem Commun 2018; 53: 12540-12543.
- Dwidar M, Yokobayashi Y. Riboswitch Signal Amplification by Controlling Plasmid Copy Number. ACS Synth Biol 2019; 8: 245-250.
- Yokobayashi Y. Applications of high-throughput sequencing to analyze and engineer ribozymes. Methods 2019; DOI: 10.1016/j.ymeth.2019.02.001.
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- Yokobayashi Y. Large-Scale Sequence-Function Analysis and Engineering of Nucleic Acid Enzymes, 8th Synthetic Bioengineering Symposium, Kobe University, Hyogo, Japan, Sep 26 (2018). Invited talk
- Yokobayashi Y. Small Molecule-Responsive Riboswitches in Mammalian Cells, 399th CBI Society Lecture Series, Tokyo Institute of Technology, Tokyo, Japan, Nov 14 (2018). Invited talk.
- Yokobayashi Y. Large-Scale Sequence-Function Relationship Analysis and Engineering of Nucleic Acid Enzymes by NGS, The 41st Annual Meeting of the Molecular Biology Society of Japan, Yokohama, Japan, Nov 30 (2018). Invited talk.
- Tabuchi T, Dwidar M, Yokobayashi Y. Selection of Aptazymes that Respond to Bioactive Ligands Using High Throughput Sequencing (HTS). The 17th Annual Bio-IT World Conference & Expo '18, Boston, U.S.A., May 15-17 (2018). Poster presentation.
- Mustafina K, Yokobayashi Y. Development of Mammalian ON-Riboswitches by High-Throughput Sequencing, The 45th International Symposium on Nucleic Acids Chemistry, Kyoto, Japan, Nov 8 (2018). Poster presentation.
- Nomura Y, Mustafina K, Rotrattanadumrong R, Yokobayashi Y. Synthetic Ribozyme Scaffold for Development of Aptazymes and Riboswitches. The 45th International Symposium on Nucleic Acids Chemistry, Kyoto, Japan, Nov 7 (2018). Poster presentation.
- Venugopal D, Nomura Y, Dwidar M, Yokobayashi Y. Photocaged Guanine Modulates Riboswitch Function by Light, The 45th International Symposium on Nucleic Acids Chemistry, Kyoto, Japan, Nov 7 (2018). Poster presentation.
5. Intellectual Property Rights and Other Specific Achievements
Provisional patent application for the development and applications of a histamine aptamer was filed.
6. Meetings and Events
6.1 Programmable Formation of Catalytic RNA Arrays and Polygons by Assembling Modular RNA Enzymes
- Date: March 7, 2019
- Venue: OIST Campus Lab3
- Speaker: Dr. Yoshiya Ikawa (University of Toyama)
7. Other
Hosted a doctroal student intern from Tokyo University of Agriculture and Technology (Kinuko Ueno) from January to March 2019.
Hosted a rotation student (Aleksandra Bliznina) from May to August 2018.
Yohei Yokobayashi was awarded a KAKENHI grant: JSPS Challenging Research (Exploratory) 18K19944 (PI), project period 6/29/2018-3/31/2020.



