2020年度
進化神経生物学ユニット
准教授 渡邉 寛

Abstract
Many of the research projects conducted in the Evolutionary Neurobiology Unit is aimed at elucidating the origins of animal systems and how key processes in their early evolution were established at the molecular and genetic levels. Specific targets of our research include the genetic and physiological features of neurons and other basic animal cells (glia, muscles, digestive cells, etc.) in the ancestral animals and early evolutionary processes of body axes and germ layers, which are fundamental to shape the multicellular animal individual.
These are biologically important issues, and although various hypotheses have been proposed over the years, there are still many unanswered questions. In our unit, we will unravel the secrets of the "birth of animals", that happened around 600 million years ago, by constructing a new systematic approach using state-of-the-art mass spectrometry, bioinformatics, and gene function analysis techniques for a group of primitive, and thus phylogenetically important, animal lineages (e.g., ctenophores and cnidarians). To further strengthen our study, we have started collaborations with marine biologists and AI researchers.
1. Staff
- Dr. Hiroshi Watanabe, Assistant Professor
- Dr. Eisuke Hayakawa, Group Leader
- Dr. Kurato Mohri, Staff Scientist
- Dr. Ryo Nakamura, Postdoctoral Scholar
- Dr. Ryotaro Nakamura, Research Unit Technician
- Dr. Yayoi Hongo, Research Unit Technician
- Chihiro Kawano, Research Unit Technician
- Akiko Tanimoto, Research Assistant
- Junko Higuchi, Research Assistant
- Hibiki Fukunaga, Research Assistant (MHLW-KAKEN)
- Kanako Hirata, Research Assistant (POC)
- Ivan Mbogo, PhD Student
- Larisa Scheloukhova, PhD Student
- Osamu Horiguchi, PhD Student (JSPS DC1 fellow)
- Christine Guzman, PhD Student (JSPS DC1 fellow)
- Kun-Lung Li, PhD Student
- Minato Miyake, PhD Student
- Chihiro Arasaki, Research Unit Administrator
Past Members
- Dr. Shinya Komoto, Staff Scientist (October 2016–March 2018) (Currently OIST Imaging Section, Staff)
- Dr. Amol Dahal, Research Unit Technician (May 2016– November 2018) (Currently Kathmandu University, Lecturer)
- Ms. Rio Zakoh, Research Assistant (April 2018 – March 2019)
- Dr. Mei-Fang Ling, Postdoctoral Scholar (May 2017-July 2020) (Currently National Sun Yat-sen University, Assistant Professor)
- Erina Kawai, Research Unit Technician (Aug 2016-Oct 2019) (Currently OIST Marine Climate Change Unit, Research Unit Scientific Diver)
2. Collaborations
2.1 Establishment of gene introduction method in marine invertebrates
- Type of collaboration: Joint research
- Researchers:
- Assist Prof. Munetsugu Bam, University of Yamanashi
- Assoc. Prof. Hiroaki Nakano, Shimoda Marine Research Center, University of Tsukuba, Shizuoka, Japan
2.2 3D structure modeling of proteins and macromoleculer complexes of basal animals
- Type of collaboration: BINDS collaboration
- Researchers:
- Dr. Kentaro Tomii (PI, Team Leader), Intelligent Bioinformatics Research Team, Artificial Intelligence Research Center (AIRC), National Institute of Advanced Industrial Science and Technology (AIST), Japan
2.3 Identification of ion channel receptors of novel neuropeptides
- Type of collaboration: Joint research
- Researchers:
- Prof. Stefan Gruender, RWTH, Aachen University, Germany
2.4 Gene function analysis in the model ctenophoran Bolinopsis mikado
- Type of collaboration: Joint research
- Researchers:
- Prof. Kazuo Inaba and Assist Prof. Kogiku Shiba, Shimoda Marine Research Center, University of Tsukuba, Shizuoka, Japan
2.5 Genome analysis of Zoantharian species
- Type of collaboration: Joint research
- Researchers:
- Assist. Prof. Mei-Fang Lin, National Sun Yat-Sen University, Taiwan
- Assoc. Prof. James Reimer, University of the Ryukyus
3. Activities and Findings
3.1 Identification and characterization of basal metazoan neuropeptides
The physiological feature (e.g. neurotransmitters) of neurons in the first animal remains unknown. Among neurotransmitter systems commonly functioning in bilaterians and cnidarians, the neuropeptide system has long been assumed to be related to the ancestral nervous system. Genome-wide surveys have been carried out in the basal animals to describe the complete set of neuropeptide genes. However, peptide prediction is still largely based on the structural features of bilaterian peptide precursors, hence providing only a few candidate genes for short neuropeptides of basal animals.
We had conducted a large scale and comprehensive mass spectrometry analysis to identify neuropeptides in basal animals including Cnidaria, Ctenophora, and Porifera. In 2020, we performed in-depth data analysis to further elucidate molecular properties and evolutionary aspects of newly identified neuropeptides. This analysis led to the identifications of cleavage sites commonly used in the Cnidaria and Ctenophora (Figure 1). Further, the precursor structures of the newly identified neuropeptides were subjected to large scale all-to-all homology search together with thousands of other neuropeptide precursors in the animal kingdom. Graph-based analysis indicated that the Ctenophora neuropeptides make unique cluster and not showing structural similarities to any other families in animal kingdom. These results illustrate that structural variation of neuropeptides had rapidly evolved, whereas the core molecular mechanisms such as precursor cleavage were strictly conserved during animal evolution.
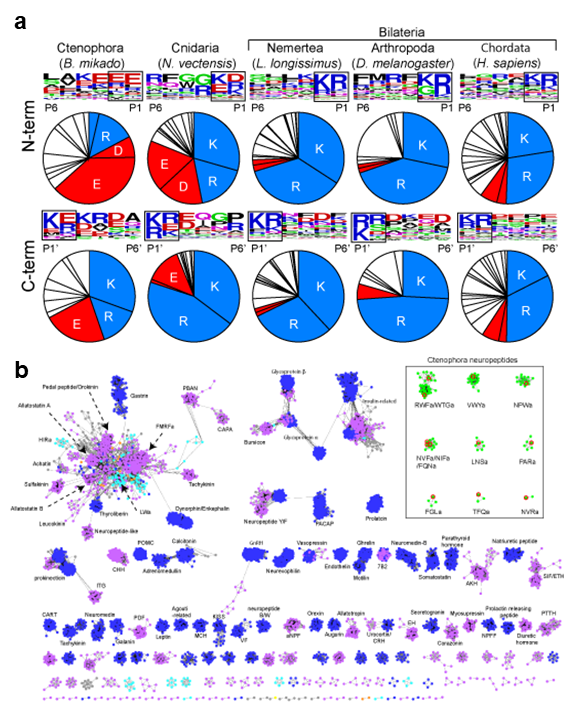
Figure 1. (a) Compositions of amino acids in the cleavage sites (2 amino acids length) and sequence LOGO map of N- and C-terminal flanking region of neuropeptides in animal kingdom. Basic amino acids (lysine and arginine) and acidic amino acids (asparatic acid and glutamic acid) are in blue and red respectively. The source of the sequences can be found in supplementary materials. (b) Cluster map of neuropeptide precursors. Edges correspond to BLAST e-value of < 1e-5. Ctenophora: red, Cnidaria: orange, Placozoa: green, Xenoacoelomorpha: gray, Protostomia: purple, Deuterostomia: blue.
3.2 Functional analysis of synaptic molecules in Cnidaria
The invention of a nervous system was a key event in evolution, allowing animals to perceive their environment and respond in appropriate ways. In the nervous system, information is exchanged between neurons via specialized structures called synapses. However, little is known about how the synaptic transmission first evolved. In this project, we focus on core and evolutionarily conserved presynaptic cell adhesion molecules (psCAMs) that have critical roles in bilaterian chemical synapse. In Cnidaria, the closest outgroup to Bilateria, psCAM homologs became fully integrated into a subset of neurons (Figure 2). We performed siRNA-mediated gene knockdown (KD) experiments to understand what the function of some psCAMs in Nematostella (Cnidaria). Gene KD of neuron-specific psCAM1 or psCAM2 didn’t affect anatomical architecture of the psCAMs/PRGa-positive neurons (not shown), but did result in a reduced rate of radial contractions, suggesting synaptic functions of these proteins. Interestingly, PRGa-KD didn’t mimic the psCAMs-KD phenotype. This may suggest that psCAM-dependent neural activity is not required for the peptide nervous system, but probably for the other neurotransmission. The synaptic function of psCAM proteins is currently being investigated in more detail.
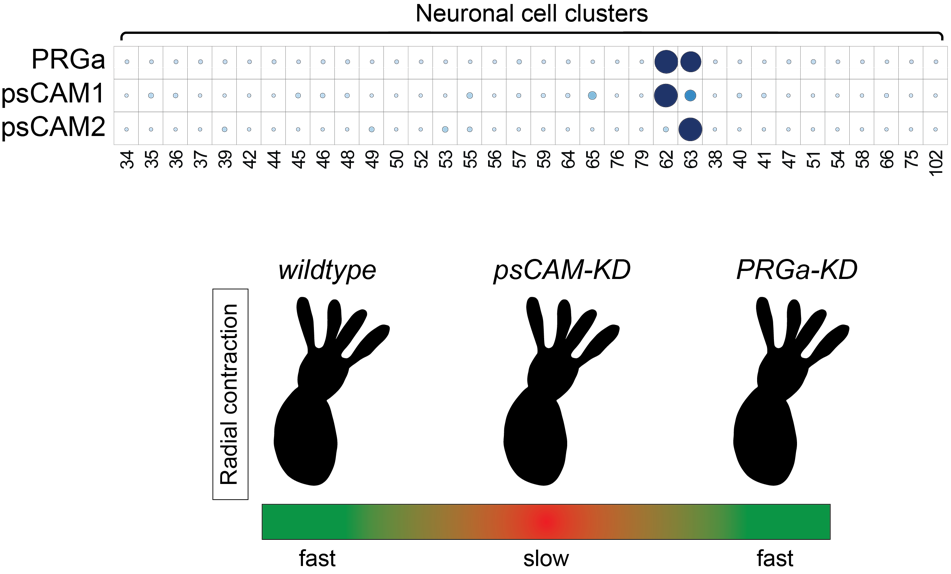
Figure 2. (top) Expression distribution of a neuropeptide (PRGamide)-precursor gene and neuronal psCAM homologs in the scRNA-seq data of adult Nematostella vectensis (Cnidaria). (bottom) Gene knockdown of psCAM1 or psCAM2 resulted to a reduced rate of radial contractions. Meanwhile, knocking down PRGamide, a neuropeptide expressed in psCAM-expressing cells (C62 and C63), did not produce the same phenotype.
3.3 Evolutionary origin(s) of neurons
The nervous system of the ctenophore, the most ancient animal, remains enigmatic. The lack of neuronal character comparable to nervous systems of other animals like Cnidaria and Bilateria has led to a debate “Is the ctenophore neuron was evolved independently?”. One efficient way to address this important biological question is to understand the neuronal charactors of ctenophore neurons, and analyze the neurogenic mechanism and comparing it with that of Cnidaria/Bilateria. To do this, we have been analyzing the function of the putative neurogenic transcription factor (TF), which is evolutionarily conserved in Cnidaria/Bilateria using a Japanese ctenophore species, Bolinopsis mikado (Figure 3). We fortunately have identified a number of neuropeptides in Ctenophora (see 3.1). Using the neuropeptides as the definitive neuronal markers, we found that one of putative neurogenic TFs is highly enriched in the neuropeptide-positive cell cluster. Knockdown of this TF led 139 and 42 genes down- and up-regulation, respectively (Fig. e-h). We now are analyzing genes to identify the function of target TF and ultimately ctenophore neurogenic mechanisms.
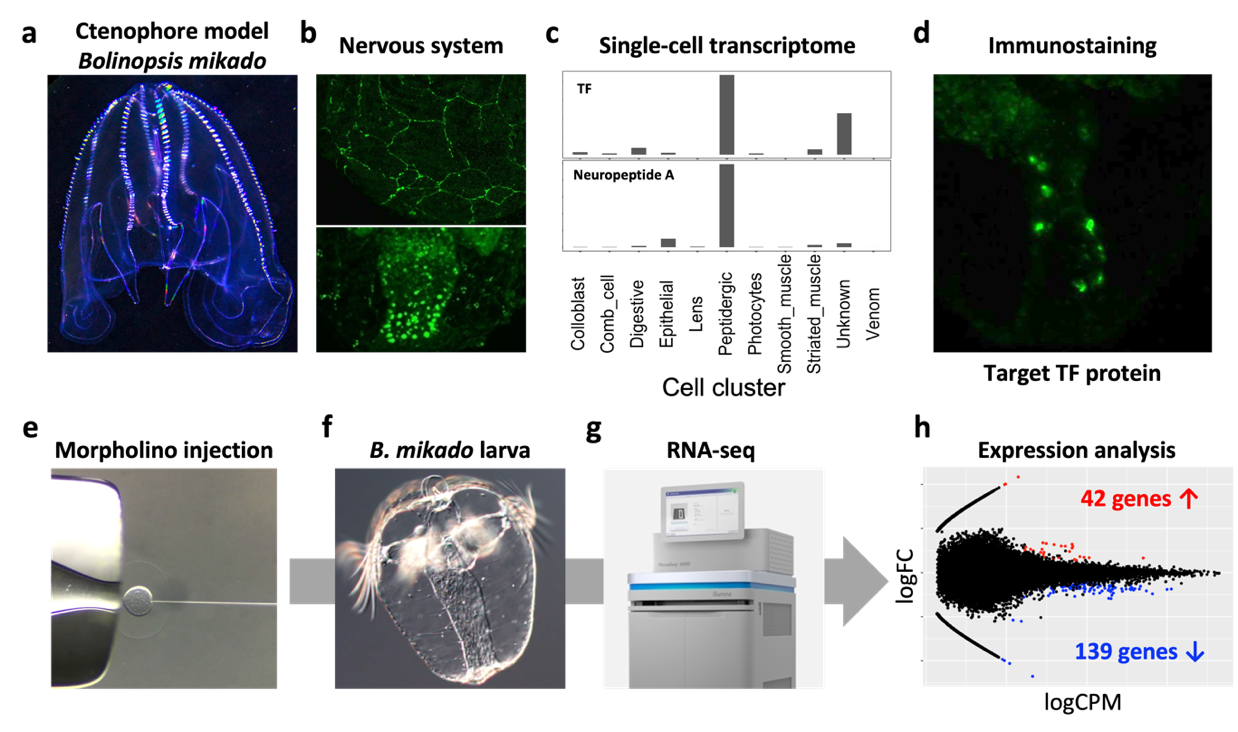
Figure 3. (a) Adult B. mikado. (b) Nervous systems of B. mikado. Epithelial nerve net (top, Neuropeptide B) and oral neuroendocrine-like cells (bottom, Neuropeptide A). (c) Single-cell expression profiles of target TF and Neuropeptide A (data derived from close species, Mnemiopsis leidyi). (d) Expression of Target TF. (e-g) our scheme of TF knowkdown by morpholino injection, RNA preparation from larvae, NovaSeq 6000 sequencing. (h) MA plot showing differentially expressed genes. Red and blue dots indicate up-and down-regulated genes, respectively. X-axis shows logFC, and Y-axis shows log CPM.
3.4 Evolutionary origins of glial cells
The nervous system of bilaterian animals is generally composed of neurons and glial cells. The evolutionary roots of glial cells remain obscure. Histological examinations have not so far revealed any morphological sign of glial cells in Cnidaria. This led to the hypothesis that glial cells appeared with the centralization of the nervous system after the common bilaterian ancestor had branched off from Cnidaria (Figure 4, left). However, this view has not been well examined at the genetic level. All the transcription factors (TFs) and signaling pathways driving gliogenesis in Bilateria are present in the genome of Cnidaria (Nematostella) (Figure 4, right). Using Nematostella as a cnidarian model organism we are exploring the functions of the conserved bilaterian gliogenic TFs in order to shed light on the evolutionary processes of glial cells.
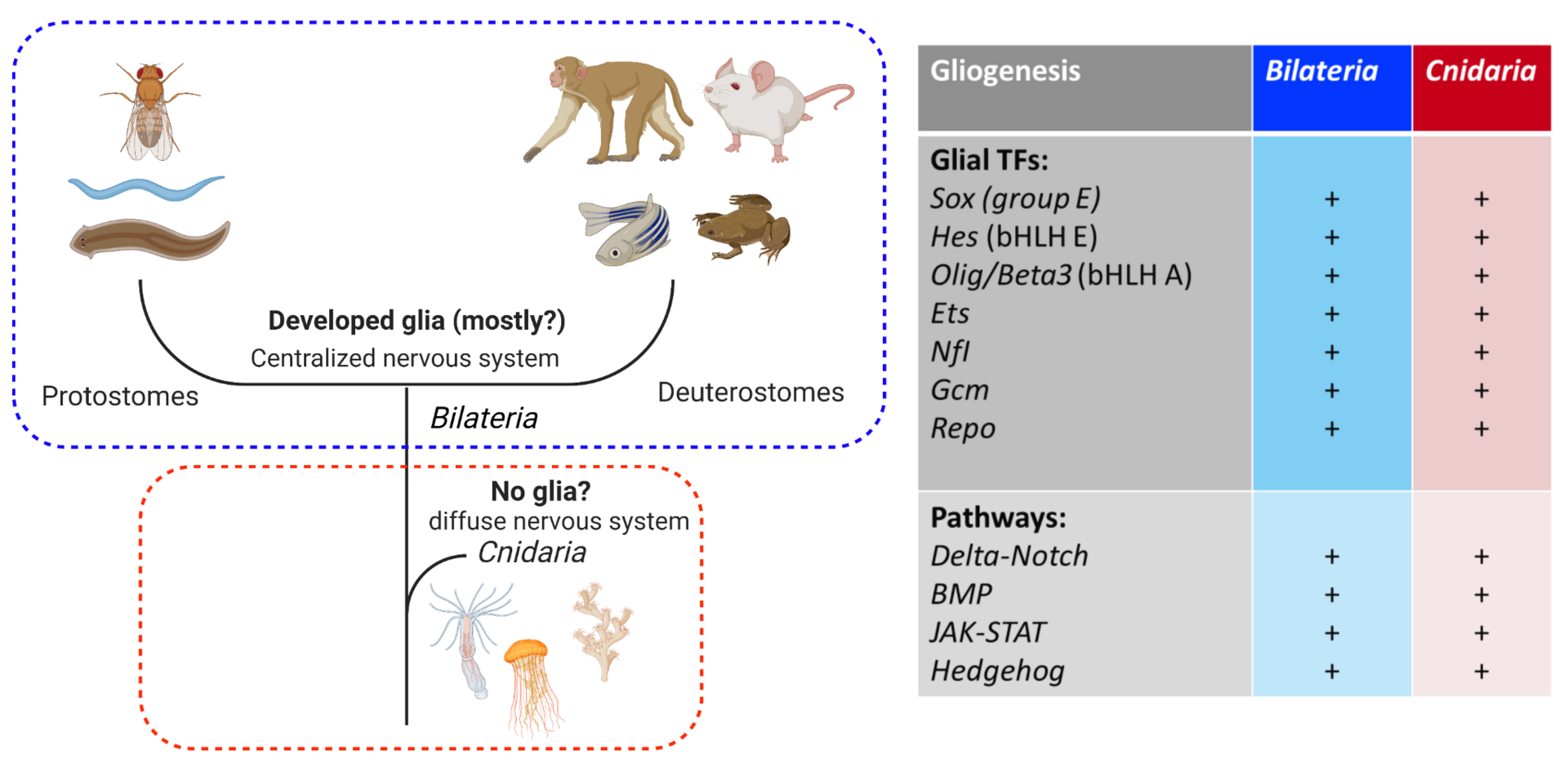
Figure 4. (Left) the current view of glial evolution based on morphological studies assumes that the centralization of the nervous system was accompanied by glial emergence. (Right) bilaterian gliogenic transcription factors and signaling pathways are conserved in Cnidaria. Cluster
3.5 Evolution of the neural centralization
It is still largely unknown whether ancestral bilaterian had a central nervous system (CNS) or not. Most of the small marine invertebrates develop a neuronal cluster around the oral/esophagus region (e.g. circumopharyngeal neurvous system: CirNS) (Figure 5), suggesting a possible architecture of the “minimum brain”. In order to examine the evolutionary conservation of genetic mechanisms required for the development of cnidarian CirNS, we performed a genome-wide analysis to characterize genes involved in the cnidarian CirNS development. In 2019, we identified 7 cnidarian transcriptional factors (TFs) which are highly conserved in bilaterian CNS, and performed a series of siRNA-mediated knockdown (KD) experiments using N.vectensis, the cnidarian model. In FY2020, we performed RNA sequencing analyses of cnidarian larvae where expression level of specific TF is reduced. We found that 49 % of downregulated differential expression genes (DEGs) are related with transcription and neuron transmission/signaling at the oral region, suggesting a requirement of the TF in the CirNS development. We are now analyzing the function of the CirNS to decipher early evolutionary process of neural centralization.

Figure 5 (a) Peptidergic neuronal assemblies related to mouth/pharynx are widely observed in Cnidaria and Bilateria, suggesting the basic and common function of the “minimal” CNS. (b) oral-specific neuron clusters which are enriched with genes related with signal transmission in bilaterian CNS. (c) Differential expression analyses in KD embryos. 114 downregulated differential expression genes (DEGs) were identified. (d) Putative functions of the down-regulated genes. (e) Downregulated DEGs were expressed in oral neuron clusters.
4. Publications
4.1 Journals
- Yi R., A. Fahrenbach A., & Y. Hongo Y. Radiolytically Driven Chemical Evolution. Journal of Geography, 129(6), 837851, 2020, doi:10.5026/jgeography.129.837
-
Atrigenio, M. P., Conaco, C., Guzman, C., Yap, H. T., & Aliño, P. M. (2020). Distribution and abundance of Heliopora coerulea (Cnidaria: Coenothecalia) and notes on its aggressive behavior against scleractinian corals: Temperature mediated? Regional Studies in Marine Science, 40. doi:10.1016/j.rsma.2020.101502
-
Mohri, K., Tanaka, R., & Nagano, S. (2020). Live cell imaging of cell movement and transdifferentiation during regeneration of an amputated multicellular body of the social amoeba Dictyostelium discoideum. Developmental Biology, 457(1), 140–149, doi: 10.1016/j.ydbio.2019.09.014
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- Sheloukhova, L. & Watanabe, H. Deciphering glial evolution: genetic and functional characterization of ancestral glia. TAGC 2020 online. The Allied Genetics Conference. April 22-25, 2020.
- Hayakawa, E., Watanabe, H., & Kondo, K. An analytical framework using multilayer mass spectral similarity network to detect unknown compounds for toxicological analysis. The 47th annual meeting of the Japanese Society of Toxicology. Online, June 29, 2020. (poster presentation)
-
Hayakawa, E., Watanabe, H., & Kondo, K. Visualization of unknown compounds in food samples using multilayer spectral network. The 26th annual meeting of the Japanese Society of Food Chemistry. Online, August 31, 2020. (poster presentation)
-
Hayakawa, E. Animal evolution indecipherable from genome : Approach from computational mass spectrometry. Workshop at the 9th Informatics In Biology, Medicine and Pharmacology. Online, September 1, 2020. (invited talk)
-
Hongo, Y.「生命と地球のこれから」Keio Astrobiology Camp 2021 (22-24, March, 2021) FY2020
-
Hongo, Y. 「Analytical chemistry for Astrobiology 生命現象をとらえる分析のチカラ」Astrobiology Seminar ONLINE, (19th, Sep. 2020)
5. Intellectual Property Rights and Other Specific Achievements
External Fundings
- Grant-in-Aid for Scientific Research (C), Japan Society for the Promotion of Science, “Reconstruction of the ancestral neurons based on the common molecular signature of peptidergic nervous systems of early branching metazoans. (ペプチド作動性神経系に共通する分子シグネチャーに基づく祖先的神経細胞の再構築)” Lead PI: Watanabe H., Co-investigator: Hayakawa E. and Inaba K. (Shimoda), Amount 4.42 M Yen, Period: Apr. 2021 – Mar. 2023
- Grant-in-Aid for Scientific Research, Ministry of Health, Labor and Welfare, ”Research on safety and risk communication for foods developed by new biotechnologies. (新たなバイオテクノロジーを用いて得られた食品の安全性確保とリスクコミュニケーションのための研究) H30-食品-一般-002)”, Lead PI: Kondo, K. (National Institute of Health Sciences)., Co-investigator: Hayakawa E., Amount 61.48 M Yen, Period: Apr. 2018 – Mar. 2022
- Grant-in-Aid for Scientific Research (C), Japan Society for the Promotion of Science," Cross-species cross-linking proteomics to elucidate the evolutionary origin of synaptic protein complexes. (横断的クロスリンクプロテオミクスによるシナプスタンパク複合体の進化的起源の解明)” Lead PI: Hayakawa, E., Amount: 4.29M Yen, Period: Apr. 2019 – Mar. 2022
- Grant-in-Aid for Scientific Research (C), Japan Society for the Promotion of Science, “Study for the reestablishment of positional information during regeneration of multicellular bodies of social amoebae. (細胞性粘菌多細胞体の再生過程における位置情報再構成機構の解析)” Lead PI: Mohri, K, Amount: 3.3 M Yen, Period: Apr. 2020 – Mar. 2023
- NINS Astrobiology Center_Astrobiology Project Research 2021, No. AB021021, Protein first-order structure originated by multi-phase heterogeneous reactions (多相不均一反応が鍵となるタンパク一次構造の起源) Lead PI: Hongo, Y. Amount: 1.6 M Yen, Period: Apr. 2020 – Mar. 2021
6. Meetings and Events
Nothing to report
7. Other
Nothing to report.



