2019年度
進化神経生物学ユニット
准教授 渡邉 寛

Abstract
In animal evolution, these soft-bodied animals branched off from our common ancestor more than 500 million years ago. Cnidaria do not have true brains, but some of them show significant level of neural condensations at certain body region. Ctenophora, one of the most basal animal lineages, have cells bearing long processes, suggesting that they have neurons. These early-branching animals are therefore very useful to study the origin and early evolutionary processes of the nervous systems.
1. Staff
- Dr. Hiroshi Watanabe, Assistant Professor
- Dr. Eisuke Hayakawa, Group Leader
- Dr. Mei Fang Lin, Postdoctoral Scholar
- Dr. Ryo Nakamura, Postdoctoral Scholar
- Erina Kawai, Research Unit Technician
- Ryotaro Nakamura, Research Unit Technician
- Chihiro Kawano, Research Unit Technician
- Yayoi Hongo, Research Unit Technician
- Akiko Tanimoto, Research Assistant
- Junko Higuchi, Research Assistant
- Hibiki Fukunaga, Research Assistant
- Kanako Hirata, Research Assistant
- Ivan Mbogo, PhD Student
- Larisa Scheloukhova, PhD Student
- Osamu Horiguchi, PhD Student
- Christine Guzman, PhD Student
- Chihiro Arasaki, Research Unit Administrator
Past Members
- Dr. Shinya Komoto, Staff Scientist (October 2016–March 2018, currently OIST Imaging Section, staff)
- Dr. Amol Dahal, Research Unit Technician (May 2016– November 2018, currently Kathmandu University, Lecturer)
- Rio Zakoh, Research Assistant (April 2018 – March 2019)
2. Collaborations
2.1 Establishment of gene introduction method in marine invertebrates
- Type of collaboration: Joint research
- Researchers:
- Dr. Munetsugu Bam, University of Yamanashi
- Prof. Hiroaki Nakano, University of Tsukuba
2.2 3D structure modeling of proteins and macromolecular complexes of basal animals
- Type of collaboration: BINDS collaboration
- Researchers:
- Dr. Kentaro Tomii, AIST (National Institute of Advanced Industrial Science and Technology)
2.3 Identification of ion channel receptors of novel neuropeptides
- Type of collaboration: Joint research
- Researchers:
- Prof. Stefan Gruender, RWTH, Aachen University, Germany
2.4 Extracellular Matrix Proteins of Cnidarians
- Type of collaboration: Joint research
- Researchers:
- Prof. Josephine C Adams, School of Biochemistry, University of Bristol, United Kingdom
- Prof. Suat Oezbek, Centre for Organismal Studies, Heidelberg University, Germany
3. Activities and Findings
3.1 Genome analysis of basal cnidarians
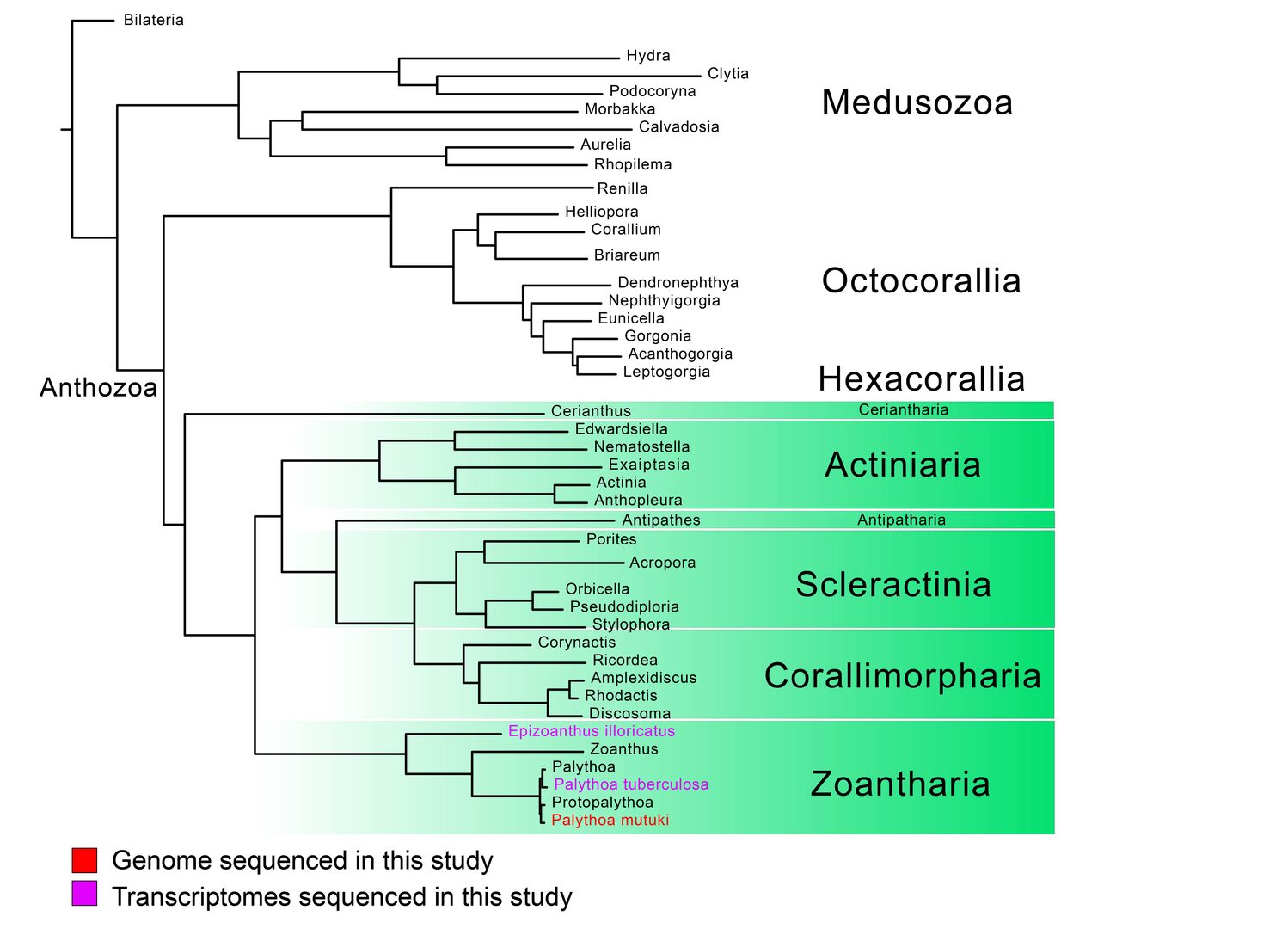
The early-branching animals such as cnidarians have possessed important evolutionary information for understanding the emergence of body plan complexity. By decoding genomes and transcriptomes of selected cnidarian species with their evolutionary importance, we aim to understand the origin of central nervous system and its primordial functions. The genome and transcriptomes of several zoantharians, which is one of the basal hexacorallians, have been decoded (Fig 1). Zoantharian larvae bear well-developed bands of cilia along their body axes that is reminiscent of ciliary bands of marine invertebrate larvae. We are interested in the cellular, physiological, and genetic features of the zoantharian nervous systems associated with their larval ciliary bands. This year our unit has deciphered genome of a zoantharian, one of the dominant zoantharian species in Okinawa water. The transcriptomes of its counterparts were also sequenced. Expending both next-generation sequencing and third-generation sequencing data, this project has provided insight into the evolution of metazoan.
3.2 Evolution of the neural centralization
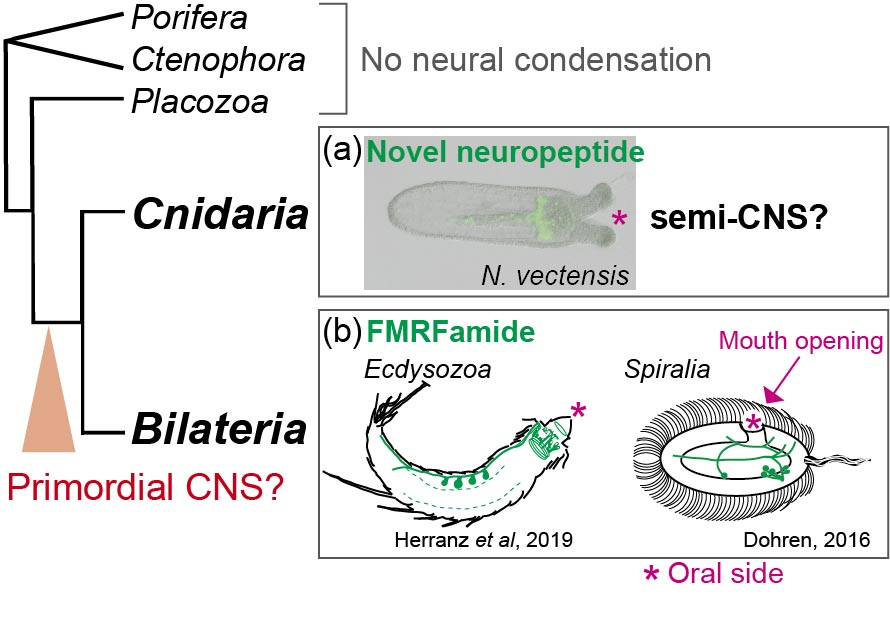
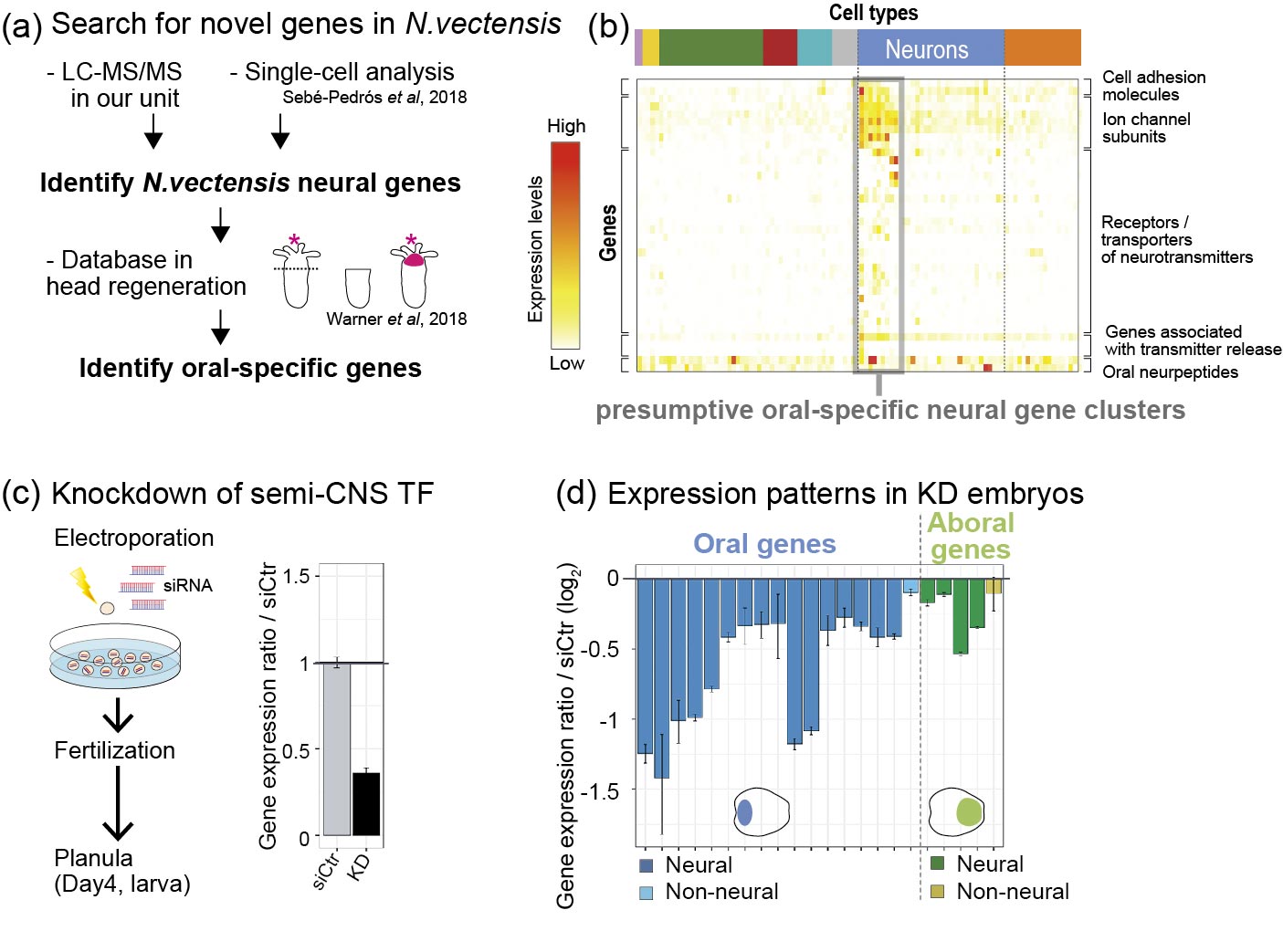
It remains unanswered whether ancestral bilaterian had a central nervous system (CNS) or not. We performed comparative genome and developmental analyses of the cnidarian semi-CNS developing at their oral region. Cnidaria is the closer sister group to all bilaterians (Fig. 2). Our analyses have so far demonstrated that the cnidarian semi-CNS comprises of 3 oral-regionalized neuropeptides and 7 bilaterian CNS-associated genes which are highly conserved in bilaterian CNS. To understand the developmental mechanism of cnidarian semi-CNS, we performed the siRNA-mediated knockdown (KD) of a semi-CNS transcription factor genes (semi-CNS TFs) in N. vectensis. (Fig. 3). We found the expressions of the semi-CNS genes are significantly downregulated by knockdown of semi-CNS TFs, implying the common neurodevelopmental mechanism is functioning in both cnidarian oral semi-CNS and bilaterian CNS.
4. Publications
4.1 Journals
- Hayakawa, E., Watanabe, H., Menschaert, G., Holstein, T. W., Baggerman, G., Schoofs, L. A combined strategy of neuropeptide prediction and tandem mass spectrometry identifies evolutionarily conserved ancient neuropeptides in the sea anemone Nematostella vectensis. PLoS One, doi: 10.1371/journal.pone.0215185 (2019).
- Hayakawa, E., Ohgidani, M., Fujimura, Y., Kanba, S., Miura, D., Kato, T. A. Cuprizone-treated mice, a possible model of schizophrenia, highlighting the simultaneous abnormalities of GABA, serine and glycine in hippocampus. Schizophr Res., doi: 10.1016/j.schres.2019.06.010 (2019).
- Guzman, C., Atrigenio M, Shinzato C, Aliño P, and Conaco C. Warm seawater temperature promotes substrate colonization by the blue coral, Heliopora coerulea. PeerJ. 7, e7785 DOI 10.7717/peerj.7785 (2019)
- Shoemark D.K., Ziegler B., Watanabe, H., Strompen J., Tucker R.P., Özbek S., Adams J.C. (2019) Emergence of a Thrombospondin Superfamily at the Origin of Metazoans. Molecular Biology and Evolution 36(6), 1220-1238, doi:10.1093/molbev/msz060
- Lin, MF., Shunichi Takahashi, Sylvain Forêt, Simon K. Davy, David J. Miller. (2019) Transcriptomic analyses highlight the likely metabolic consequences of colonization of a cnidarian host by native or non-native Symbiodinium species. Biology Open 8: bio038281 doi: 10.1242/bio.038281
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- Guzman, C., Tomii, K., Watanabe, H. Functional and Structural Analyses of Neurexin-Mediated Synaptic Complexes in Early-Branching Animals. The Identity and Evolution of Cell Types. EMBL, Heidelberg, Germany, May 15–18, 2019 (2019)
- Hayakawa E. Omics data analysis. Workshop hosted by the division of Bio analysis of the society for biotechnology Japan. Miyazaki, Japan, July 26, 2019 (2019) (invited talk)
- Hayakawa, E. False discovery rate in peptidomics, The 3rd Hackathon for Mass Spectrometry Informatics, Kagoshima, Japan, July 7–12, 2019 (2019)(Invited talk)
- Lin, M.F., Reimer J., Watanabe H. Cnidarian phylogenomics to understand early evolution of Bilateria. Poster Presentation, The 67th NIBB Conference / The 6th Quest for Orthologs Meeting, July 31–August 2, 2019 (2019)
- Nakamura, R., Nakamura, R., Komoto, S., Guzman, C., Hayakawa, E., Watanabe, H. Neural centralization in early animal evolution; lessons from cnidarian studies. Oral presentation, 21stof Annual Meeting of the Society of Evolutionary Studies, Japan, Hokkaido, Japan, August 8, 2019 (2019)
- Horiguchi, O., Lin, M. F., Guzman, C., Hayakawa, E., Shigenobu, S., Jokura, K., Shiba, K., Inaba, K., Watanabe, H. Identification of Neurogenic bHLH in ctenophore Bolinopsis mikado, 21st of Annual Meeting of the Society of Evolutionary Studies, Japan, Hokkaido, Japan, August 8, 2019, (2019)
- Mbogo, I., Yasuoka.Y., & Watanabe, H. Proteomic Approach to Dissect Ancestral β-Catenin Transcription Machinery. 21st of Annual Meeting of the Society of Evolutionary Studies, Japan, Hokkaido, Japan, August 8, 2019 (2019)
- Nakamura, R., Nakamura, R., Komoto, S., Guzman, C., Hayakawa, E., Watanabe, H. 刺胞動物准中枢神経系の発生メカニズムから中枢神経系の起源に迫る, 90thAnnual Meeting of the Zoological Society of Japan, Osaka, Japan, September 2019 (2019)
- Hayakawa, E., Horiguchi, O., Kawano, C., Jokura, K., Shiba, K., Inaba, K., Watanabe, H. Mass Spectrometry-based neuropeptidomics to dissect Ctenophore nervous system, Hydra Meeting, Tutzing, Germany, September 16–19, 2019 (2019)
- Kawano, C., Hayakawa, E., and Watanabe, H. Combining Tandem Mass Spectrometry and High-Throughput peptide Synthesis for High-Confidence Neuropeptidomics. The 56th Japanese Peptide Symposium. Tokyo, Japan, October 23–25, 2019 (2019)
5. Intellectual Property Rights and Other Specific Achievements
External Fundings
-
Grant-in-Aid for Scientific Research, Ministry of Health, Labor and Welfare, ”Research on safety and risk communication for foods developed by new biotechnologies. (新たなバイオテクノロジーを用いて得られた食品の安全性確保とリスクコミュニケーションのための研究)H30-食品-一般-002)”
- Lead PI: Kondo K (National Institute of Health Sciences).
- Co-investigator: Hayakawa E (OIST)
- Amount 61.48 M Yen
- Period: Apr. 2018 – Mar. 2022
-
Grant-in-Aid for Scientific Research (C), Japan Society for the Promotion of Science," Cross-species cross-linking proteomics to elucidate the evolutionary origin of synaptic protein complexes. (横断的クロスリンクプロテオミクスによるシナプスタンパク複合体の進化的起源の解明)”
- Lead PI: Hayakawa, E
- Amount: 4.29M Yen
- Period: Apr. 2019 – Mar. 2022
6. Meetings and Events
Nothing to report
7. Other
Nothing to report.



