Octopus Genome
This is a preliminary page (in progress) that contains basic data used in the analysis of the Octopus bimaculoides genome [link to the paper]. Please see below for contact information, if you have any questions or require data not listed here.
- Genome assembly, annotation
- Browsers
- Whole genome/transcriptome phylogeny files
- Global gene family expansion analysis
- Repeat annotation
- Cephalopod novelties table
- Synteny dynamics and simulations
- RNA editing
- Other
- Contact
1. Genome assembly, annotation
Also available at: http://genome.jgi.doe.gov/pages/dynamicOrganismDownload.jsf?organism=Metazome
- Genome FASTA: [link]
- GFF3 gene model locations: [link]
- Protein sequences: [link]
- CDS sequences: [link]
- Annotations: PFAM / Panther / GO
- Mapping between OBI* and Ocbi* identifiers: [link] (OBI* identifiers are used for all subsequent analyses)
3. Whole genome/transcriptome phylogeny files
- Download link
- (contains all 1-1 gene families used in the alignment, their alignments, g-block results as well as concatenated alignments)
4. Global gene family expansion analysis
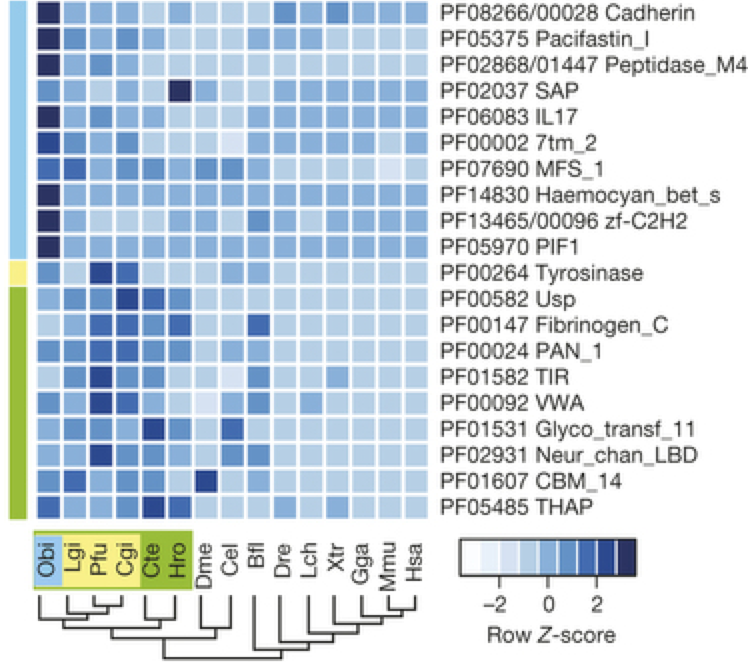
- Download raw counts table and the R script: [link]
- Repeat annotation was done using curated RepeatScout and RepeatModeller libraries:
- Main library: [link]
- Main masking gff file: [link]
- Main masking out file: [link]
- RepeatModeller only library: [link]
- Download Excel file with link information: [link]
7. Synteny dynamics and simulations
- Syntenic blocks detected among all species in the paper: [link]
- Species distribution table for each block: [link]
- Loss of pairwise synteny in Octopus compared to core conserved syntenic blocks between Capitella and Lottia. Species from left to right: Octopus, Lottia, Capitella, Helobdella. Individual scaffolds are plotted as grey rectangles, size scaled to the total length of the represented scaffolds.

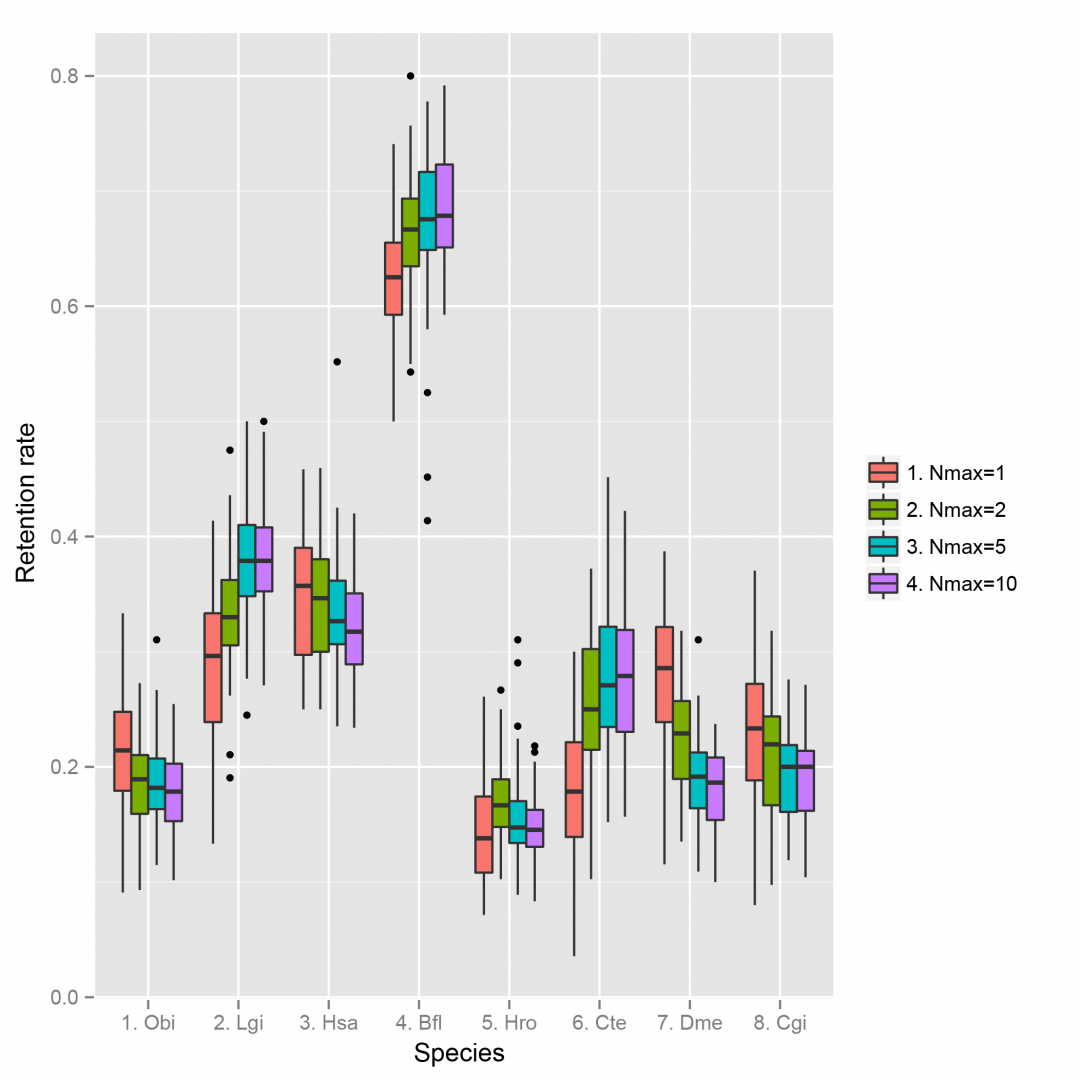
- Retention rate of bilaterian linkages across different species using different Nmax (maximum allowed intervening genes) parameter and using simulated same-length (number of genes) scaffolds as described in the paper.
Extensive RNA editing has been described in squid. To explore potential RNA editing in octopus, we mapped RNA-seq reads to the genome with TopHat (Trapnell et al. 2012). SAMtools and bcftools (Li and Durbin 2009) were used to identify differences between the genome and the transcriptome. Polymorphic positions identified in the genome were removed. The detected DNA-RNA differences for each transcriptome are listed in separate sheets within each excel document. One excel sheet contains the unfiltered results (DNA_RNA_differences_transcriptomes.xlsx), while the other only contains sites with a phred score of 40 or more (99.99% accuracy; DNA_RNA_differences_transcriptomes_phred40.xlsx). The tissues for the genome and the transcriptomes were isolated from different animals, so the identified differences represent candidate editing sites as well as polymorphisms and errors.
- Octopus has a bimodal intron size distribution [link]
- Questions: oleg.simakov [at] oist.jp



