FY2022 Annual Report
Evolutionary Neurobiology Unit
Assistant Professor Hiroshi Watanabe

Abstract
The Evolutionary Neurobiology Unit aims to elucidate the evolutionary origin of complex animal systems composed of nerves, muscles, digestive cells, etc., and the processes in their early evolution at the molecular level. Although these are biologically important questions and various hypotheses have been proposed over the decades, there are still many open questions. Our unit conducts analyses of extant species of early-branching and therefore phylogenetically important animal lineages (e.g., ctenophores and cnidarians). We are also trying to unravel the mystery of the early animal evolution by constructing a new systematic EvoDevo approach that makes full use of cutting-edge mass spectrometry, bioinformatics, gene function analysis, and machine learning technologies. We are collaborating with laboratories in various research fields with cutting-edge analytical techniques to further strengthen our interdisciplinary research.
1. Staff
- Dr. Hiroshi Watanabe, Assistant Professor
- Dr. Eisuke Hayakawa, Group Leader
- Dr. Kurato Mohri, Staff Scientist
- Dr. Ryo Nakamura, Postdoctoral Scholar
- Dr. Hongdi Wang, Postdoctoral Scholar
- Dr. Ryotaro Nakamura, Research Unit Technician
- Dr. Yayoi Hongo, Research Unit Technician
- Chihiro Kawano, Research Unit Technician
- Kanako Hirata, Research Unit Technician (PoC)
- Mika Ogata, Research Assistant (PoC)
- Asami Kyan, Research Assistant (PoC)
- Osamu Horiguchi, PhD Student (JSPS DC1 fellow)
- Christine Guzman, PhD Student (JSPS DC1 fellow)
- Minato Miyake, PhD Student
- Sen Hedife, PhD Student
- Daniel Soto Carballo, Internship Student (May-November 2022)
- Chihiro Arasaki, Research Unit Administrator
-
Past members
- Ivan Mbogo, PhD Student (September 2015–March 2022. Currently Associate of Hematology Business Development, Sysmex Corporation)
- Larisa Sheloukhova, PhD Student (September 2015–March 2022. Currently Program-specific researcher at SACI, Kyoto University)
- Kun-Lung Li, PhD Student (September 2016–September 2021. Currently PostDoc, Institute of Cellular and Organismic Biology, Academia Sinica, Taiwan)
- Junko Higuchi, Research Assistant (June 2019–May 2022)
- Akiko Tanimoto, Research Assistant (February 2019–February 2022)
- Hibiki Fukunaga, Research Assistant (October 2019–March 2021)
- Rio Zakoh, Research Assistant (April 2018–March 2019)
- Dr. Mei-Fang Lin, Postdoctoral Scholar (May 2017–July 2020. Currently Assistant Professor, National Sun Yat-sen University, Taiwan)
- Ms. Erina Kawai, Research Unit Technician (August 2016–October 2019. Currently Research Unit Scientific Diver, OIST Marine Climate Change Unit)
- Dr. Amol Dahal, Research Unit Technician (May 2016–November 2018. Currently Assistant Professor, Kathmandu University)
- Dr. Shinya Komoto, Staff Scientist (October 2016–March 2018. Currently Research Support Specialist, OIST Scientific Imaging Section)
2. Collaborations
2.1 Gene function analysis in the model ctenophoran Bolinopsis mikado
- Type of collaboration: Joint research
- Researchers:
- Professor Kazuo Inaba and Assistant Professor Kogiku Shiba, Shimoda Marine Research Center, University of Tsukuba, Shizuoka, Japan
2.2 Genome analysis of Zoantharian species
- Type of collaboration: Joint research
- Researchers:
- Assistant Professor Mei-Fang Lin, National Sun Yat-sen University, Taiwan
- Associate Professor James Reimer, University of the Ryukyus
2.3 AI-based peptide receptor prediction
- Type of collaboration: Joint research
- Researchers:
- Dr. Honoo Satake and Dr. Akira Shiraishi, Bioorganic Research Institute, Suntroy Foundation for Life Sciences, Kyoto, Japan
3. Activities and Findings
3.1 Analysis of neurotransmitters in basal metazoans
Chemical neurotransmitters play essential roles in the bilaterian nervous systems. Some of primitive neurotransmitters/modulators, including neuropeptides, have been reported to be functioning in Cnidaria, the closest sister to all bilaterians, suggesting that the last common Cnidaria/Bilateria ancestor had already equipped with the chemical neurotransmission. However, the molecular repertory of neurotransmitters has been inconclusive in Cnidaria due to the conflicting results obtained by different techniques. Additionally, new analytical approach has been required to identify the molecules distinguishing from the numerous structural analogues. In order to separate accumulated dietary intakes from the endogenous synthetic products, we surveyed mass-shifted molecules in the LC-MS metabolomics applying stable isotope labeled precursors to the animals. Furthermore, the identified molecule was subjected to imaging MALDI to visualize the location. These techniques largely improved the precision of endogenous transmitter identification and their tissue distribution, comparing to the less-specific staining with conventional immunohistochemistry, and enabled us to consider the local function of the molecules.
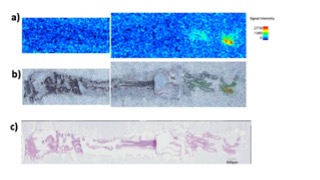
Figure 1: (a) MALDI Ion imaging of one of chemical transmitter (obtained by MIRUION, Osaka Japan). (b) Merged image of the ion image (a) and microscopic image (c). (c) H&E staining.
3.2 Evolutionary origin(s) of the central nervous systems
The evolutionary origins of the central nervous systems (CNSs) are still largely unknown (Figure 2a). Homologs of transcription factors (TFs) involved in CNS patterning in Bilateria exists also in cnidarian genomes, but their developmental and physiological functions have not so far been clarified. A comprehensive genome surveys and functional analyses using Nematostella vectensis (Cnidaria) revealed that the genetic features of the cnidarian oral/pharyngeal nervous system (PhNS) share molecular similarities with the bilaterian CNSs. In this year, we generated PhNS-deficient N. vectensis to reveal behaviors involving PhNS function (Figure 2b). A series of behavioral experiments identified neural assemblage expressing neuropeptides and TFs that regulate broad range of mouth/pharynx-related systematic responses in N. vectensis (Figure 2c). Our results suggest that some behavioral and metabolic functions observed in bilaterian CNS were already controlled by functionally localized neural assemblies even in the diffuse nervous system prior to emergence of bilaterian CNSs.
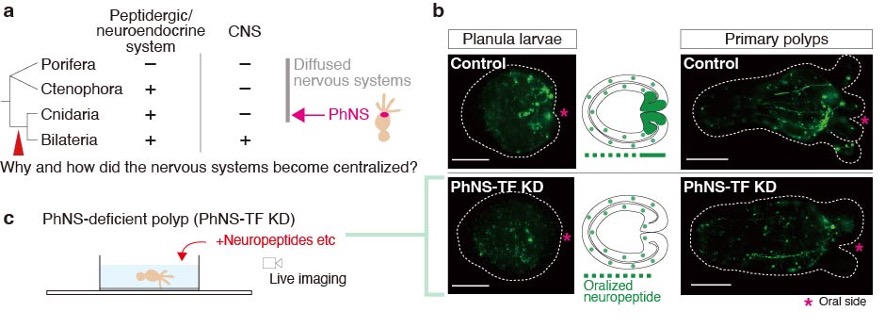
Figure 2: (a) A phylogenetic relationship of metazoan lineages and CNS evolution. (b) Development of PhNS peptidergic neurons in control and CNS-TF-depleted larvae and polyps of N. vectensis. Scale bars, 100 µm. (c) A schematic illustration of behavioral analysis using PhNS-deficient polyps.
3.3 Neurogenesis of ctenophore neurons
Animals have acquired animal-specific cell types such as neurons, in their early evolutionary phase, but their evolutionary origin remains enigmatic. Ctenophora are the earliest divergent extant animal phylum bearing neurons and therefore they provide an important information for understanding the evolutionary origin of neurons. In this study, we sought to identify in Bolinopsis mikado (Ctenophora) transcription factors (TFs) that is evolutionarily conserved in Ctenophora/Cnidaria/Bilateria. We have identified TFs that are expressed in the cells with neuropeptides. Knockdown of one of the TFs resulted in expression changes of 162 genes (39 up, 123 down) (Figure 3a). We analyzed the expression patterns of these genes in Mnemiopsis leidyi (Ctenophora) (Figure 3b). Visualization of two down-regulated genes, which are highly expressed in neuropeptide-expressing cells, revealed that were expressed in cells at pharynx (Figure 3c, d).
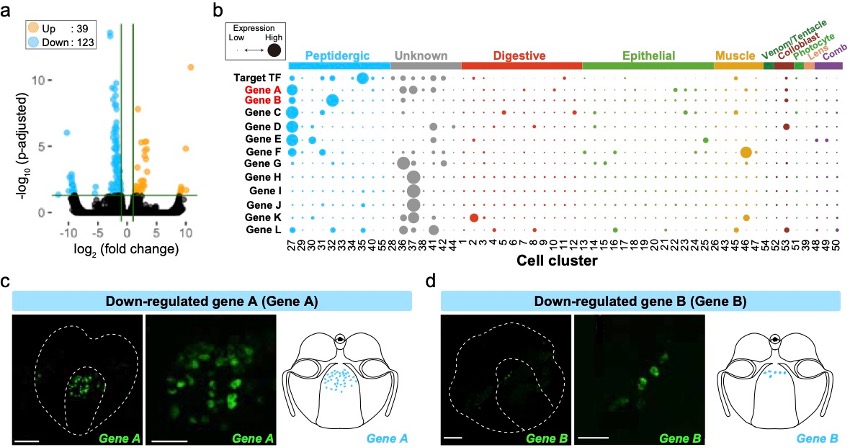
Figure 3: (a) Volcano plot of the RNA-seq analysis comparing with control and target TF knocked down. Orange and blue dots indicate up- and down-regulated genes, respectively. (b) Dot plot of target TF and down-regulated genes in M. leidyi scRNA-seq. The dot size indicates the expression level, and the dot color indicates the cell type. (c, d) Fluorescent in situ hybridization images and schematics of the down-regulated genes A (c) and B (d). Scale bars for the whole-body image and zoomed image are 50 µm and 20 µm, respectively.
4. Publications
4.1 Journals
- Rossetto, D., Valer, L., Yeh Martín, N., Guella, G., Hongo, Y., Mansy, S. S., Prebiotic Environments with Mg2+ and Thiophilic Metal Ions Increase the Thermal Stability of Cysteine and Non-cysteine Peptides. ACS Earth and Space Chemistry, 6 (5), 1221-1226, doi:10.1021/acsearthspacechem.2c00042 (2022)
- Hayakawa, E., Guzman, C., Horiguchi, O., Kawano, C., Shiraishi, A., Mohri, K., Lin, M. F., Nakamura, R., Nakamura, R., Kawai, E., Komoto, S., Jokura, K., Shiba, K., Shigenobu, S., Satake, H., Inaba, K., & Watanabe, H. Mass spectrometry of short peptides reveals common features of metazoan peptidergic neurons. Nature Ecology & Evolution, 6 (10), 1438-1448, doi: 10.1038/s41559-022-01835-7 (2022)
- Hongo, Y., Sekimoto, K., Acquiring MS Data. Journal of the Mass Spectrometry Society of Japan, 70 (3) 202-208, doi:10.5702/massspec.S22-53 (2022)
- Hongo, Y., Sekimoto, K., Interpretation of Mass Spectra_1. Journal of the Mass Spectrometry Society of Japan, 70 (4), 274-279, doi: 10.5702/massspec.S22-64 (2022)
- Masuda-Ozawa, T., Fujita, S., Nakamura, R., Watanabe, H., Kuranaga, E., & Nakajima, Y. siRNA-mediated gene knockdown via electroporation in hydrozoan jellyfish embryos. Scientific Reports, 12 (16049), doi:10.1038/s41598-022-20476-1 (2022)
4.2 Books and other one-time publications
- Guzman, C., Shedding light on the elusive neurons of comb jellies. Nature Ecology & Evolution, August 9, 2022 (2022)
4.3 Oral and Poster Presentations
- Hayakawa, E., Watanabe, H., Computational mass spectrometry to analyze unknown compounds in environmental samples, Joint Conference on Environmental Chemicals, June 14–16, 2022, Toyama, Japan (Oral Presentation)
- Hayakawa, E., Guzman, C., Horiguchi, O., Kawano, C., Nakamura, R., Inaba, K., Watanabe, H., Peptidomics and single cell transcriptome reveal the evolutional origin of nervous system, The 70th Annual Conference on Mass Spectrometry, Japan, June 22–24, Fukuoka, Japan (Oral Presentation)
- Watanabe, H. Common Features of Metazoan Peptidergic Neurons Support Their Single Origin. West Pacific Marine Biology International Conference, Online, June 27, 2022 (Oral Presentation)
- Horiguchi, O., クシクラゲのペプチド発現神経の特徴と機能から見る神経細胞の進化的起源, The 24th Annual Meeting of Society of Evolutionary Studies, Japan, August 4–7, 2022, Shizuoka, Japan (Oral Presentation)
- Horiguchi, O. Characteristics and evolution of peptide expressing neuron in Ctenophora. Invited Seminar at Shimoda Marine Research Center, University of Tsukuba, August 9, Shizuoka, Japan (Oral Presentation)
- Watanabe, H., et al., Evolutionary origin of neurons: insights from peptide-expressing neurons of ctenophores. 「クシクラゲのペプチド発現神経の特徴と機能から見える神経細胞の進化的起源」, The 93rd Annual Meeting of the Zoological Society of Japan, September 8, 2022, Tokyo, Japan (Oral Presentation)
- Mohri, K., Horiguchi, O., Hayakawa, E., Watanabe, H. Conservation of the peptidergic nervous system through the Ctenophora species, The 93rd Annual Meeting of the Zoological Society of Japan, September 10, 2022, Tokyo, Japan (Oral Presentation)
- Miyake, M., Watanabe, H., Insights in evolutionary origins of the nervous system from Ctenophora Pou transcription factor functions. 「クシクラゲ Pou 転写因子群の機能解析で迫る神経の進化的起源」, The 93rd Annual Meeting of the Zoological Society of Japan, September 10, 2022, Tokyo, Japan (Oral Presentation)
- Hayakawa, E., Guzman, C., Horiguchi, O., Kawano, C., Nakamura, R., Inaba, K., Watanabe, H., ペプチドミクスとシングルセルトランスクリプトミクスが解き明かすペプチド作動性神経の進化的起源, The 16th Metabolome Symposium, September 14–16, 2022, Yamagata, Japan (Oral Presentation)
- Hayakawa, E., Watanabe, H., Kondo, K. 質量分析インフォマティクスによる 食品試料の迅速・ノンターゲットな未知化合物解析プラットフォーム, The 118th Conference on Japanese Society for Food Hygiene and Safety, November 10–11, 2022, Nagasaki, Japan (Poster Presentation)
- Hongo, Y. 質量分析とメタボロミクスが蜜月である所以, Mass Spectrometry Seminar, OoP Net, December 2, 2022, University of the Ryukyus, Okinawa, Japan (Oral Presentation)
- Guzman, C., et al., Mass spectrometry of short peptides reveals common features of metazoan peptidergic neurons, The 45th Annual Meeting of the Molecular Biology Society of Japan (MBSJ2022), November 30–December 2, 2022, Chiba, Japan (Oral Presentation)
5. Intellectual Property Rights and Other Specific Achievements
External Fundings
- Grant-in-Aid for Scientific Research (C), Japan Society for the Promotion of Science, “Reconstruction of the ancestral neurons based on the common molecular signature of peptidergic nervous systems of early branching metazoans. (ペプチド作動性神経系に共通する分子シグネチャーに基づく祖先的神経細胞の再構築)” Lead PI: Watanabe H., Co-investigator: Hayakawa E. and Inaba K. (Shimoda), Amount 4.42 M Yen, Period: Apr. 2020– Mar. 2023
- Grant-in-Aid for Scientific Research, Ministry of Health, Labor and Welfare, ”Research on safety and risk communication for foods developed by new biotechnologies. (新たなバイオテクノロジーを用いて得られた食品の安全性確保とリスクコミュニケーションのための研究), Lead PI: Kondo, K. (National Institute of Health Sciences)., Co-investigator: Hayakawa E., Amount 30.13 M Yen, Period: Apr. 2021 – Mar. 2024
- Grant-in-Aid for Scientific Research (C), Japan Society for the Promotion of Science," Cross-species cross-linking proteomics to elucidate the evolutionary origin of synaptic protein complexes. (横断的クロスリンクプロテオミクスによるシナプスタンパク複合体の進化的起源の解明)” Lead PI: Hayakawa, E., Amount: 4.29M Yen, Period: Apr. 2019 – Mar. 2023
- Grant-in-Aid for Scientific Research (C), Japan Society for the Promotion of Science, “Study for the reestablishment of positional information during regeneration of multicellular bodies of social amoebae. (細胞性粘菌多細胞体の再生過程における位置情報再構成機構の解析)” Lead PI: Mohri, K, Amount: 4.29 M Yen, Period: Apr. 2020 – Mar. 2023
6. Meetings and Events
Nothing to report
7. Other
Nothing to report.



