FY2020 Annual Report
Developmental Neurobiology Unit
Professor Ichiro Masai

Abstract
Developmental neurobiology unit uses zebrafish as an animal model and will elucidate genetic program that regulate eye development. Specific research projects are to elucidate:
(1) Mechanisms that regulate retinal cell differentiation and neural circuit formation
(2) Mechanisms that regulate photoreceptor degeneration
(3) Mechanisms that regulate lens fiber differentiation
During vertebrate development, the retina is originally derived from anterior neural plate. In this region, six major classes of retinal neurons differentiate and form neural circuits responsible for vision. Thus, the retina provides a good model for studying cell differentiation and neural circuit formation in the developing brain. First, we will investigate mechanisms that regulate cell differentiation and neural circuit formation. Second, we will investigate mechanisms that regulate photoreceptor degeneration. Photoreceptor degeneration is an important topic for medical research, because more than 300 genetic mutations are associated with photoreceptor degeneration in humans. We focus on zebrafish mutants, in which photoreceptor degeneration is caused by defects in protein synthesis in ER, protein transport from ER to the apical photoreceptive membrane region, and phototransduction. Through these mutant analyses, we will determine how photoreceptors monitor abnormalities in cellular functions and trigger apoptosis. We will also investigate the role of microglia in photoreceptor degeneration. Third, we will investigate lens development. The lens is an intraocular organ that focuses visual image on retinal photoreceptors. Lenses consist of two cell-types: lens epithelial cells and lens fiber cells. Lens epithelial cells differentiate into lens fiber cells, each of which are integrated in a stereotyped geometric pattern to form spherical lens fiber core. We will investigate mechanisms that regulate lens fiber differentiation. Through these projects, we will establish key concepts that govern development of multicellular organisms and also contribute to our understanding of pathological processes of human retinal diseases.
1. Staff
- Dr. Yuko Nishiwaki, Group leader
- Dr. Yuki Sugiyama, Staff scientist
- Dr. Yuki Takeuchi, Staff scientist
- Dr. Wei-Chieh Chiang, Staff scientist
- Dr. Luis Carretio, Postdoctral scholar (HFSP-> PEREX on 1st August)
- Dr. Nishtha Ranawat, phD student -> Junior Research Fellow (–Dec 2020)
- Ms. Manana Kutsia, phD student
- Ms. Mai Omar Abdulrahman Ahmad, phD student
- Ms. Swathy Babu, phD student
- Mr. Bedish Chatterjee, phD student
- Mr. Hung-Ju Chiang, phD student
- Mr. Dongpeng Hu, phD student
- Ms. Darshini Ravishanker, phD student
- Ms. Sarianna Touvinen, phD student (Sept 2020–)
- Mr. Yutaka Kojima, Technician
- Mr. Kevin Jeff Liner, Technician
- Dr. Tetsuya Harakuni, Technician
- Dr. Mamoru Fujiwara, Technician (Tempo staff–>PEREX on 1st October)
- Mr. Takuya Kamichika (Tempo staff, HFSP->OPEX on 1st August)
- Ms. Versha Venkatesha Murthy, Technician
- Ms. Moe Inafuku, Laboratory Assistant
- Ms. Chitose Mizuta, Laboratory Assistant
- Ms. Rui Inoue, Laboratory Assistant
- Ms. Fujino Ishibashi, Laboratory Assistant
- Ms. Madoka Makiya, Research Assistant (Tempo staff)
- Ms. Miki Kitamura, Research Assistant (Tempo staff)
- Ms. Mizuki Otake, Research Administrator/Secretary
2. Collaborations
2.1 The Title or Name or Topic of the Collaboration
- Description:In vivo functional analysis of hypoxia response genes using the zebrafish retina
- Type of collaboration: Joint research agreement
- Researchers:
- Dr. Ichiro Masai (Developmental neurobiology unit, OIST)
- Dr. Masayuki Matsushita (Department of Medicine, Ryukyu University)
2.2 The Title or Name or Topic of the Collaboration
- Description:Role of zebrafish BANP protein in tumor suppression of melanoma
- Type of collaboration: Joint research agreement
- Researchers:
- Dr. Ichiro Masai (Developmental neurobiology unit, OIST)
- Dr. Yutaka Kikuchi (Department of Biological Science, Hiroshima University)
3. Activities and Findings
3.1 Mechanism underlying photoreceptor degeneration
Apoptosis is observed in developing tissues and is believed to remove abnormal cells. Although apoptosis is important for establishment of proper neural circuits by eliminating abnormally differentiated neurons, it is unclear how differentiating cells monitor their own abnormality, and how the threshold at which apoptosis is induced is determined. Photoreceptors provide a useful model for studying such a surveillance mechanism of neuronal development and homeostasis, because there are many hereditary retinal diseases in humans associated with photoreceptor degeneration. Although 271 genes linked to hereditary retinal diseases have already been identified, these genes encode diverse functions, including phototransduction, retinol metabolism, and intracellular protein transport. To answer how photoreceptors monitor their differentiation status and homeostasis, and what kinds of molecular network determine the choice between cell survival and cell death in photoreceptors, we have investigated zebrafish mutants in which photoreceptor degeneration is triggered by defects in phototransduction, intracellular vesicular transport, and intraflagellar transport via the primary cilium. We also investigated the role of brain-resident immune cells, microglia, in photoreceptor degeneration.
3.1.1 Mechanism of photoreceptor degeneration in response to vesicular fusion defects
Intracellular protein transport is mediated by transport vesicles and its defects are often linked to photoreceptor degeneration in humans. However, it is uncertain how vesicular transport defects cause photoreceptor degeneration. We isolated zebrafish β-soluble N-ethylmaleimide-sensitive factor attachment protein (β-SNAP) mutants, in which photoreceptors differentiate, but undergo apoptosis prior to maturation (Nishiwaki et al., 2013) (Fig. 1A). β-SNAP cooperates with N-ethylmaleimide-sensitive factor (NSF) to promote recycling of SNAP receptors (SNARE), a key component of vesicular fusion machinery, by disassembling the cis-SNARE complex generated in the vesicular fusion process (Jahn and Scheller, 2006). Because it is likely that all combinations of cis-SNARE complexes fail to be disassembled in β-SNAP mutants, the mutant provides a good model for studying how vesicular transport defects cause apoptosis in photoreceptors. We found that photoreceptor apoptosis in β-SNAP mutants depends on one of t-SNARE proteins, BNip1. BNip1 normally regulates retrograde transport from Golgi to ER as a t-SNARE component of the syntaxin-18 complex. Interestingly, BNip1 has the BH3 domain, which activates apoptosis via modulation of apoptotic regulators Bax and Bcl2. We found that loss of β-SNAP induces accumulation of syntaxin-18
cis-SNARE complex, in which the BNip1 BH3 domain is activated to induce apoptosis. Thus, the syntaxin-18 cis-SNARE complex functions as an alarm factor that monitors vesicular fusion competence, and BNip1 transforms vesicular fusion defects into photoreceptor apoptosis (Fig. 1B) (Nishiwaki et al., 2013). To our knowledge, BNip1 is the first molecule that directly links vesicular fusion defects and apoptosis.
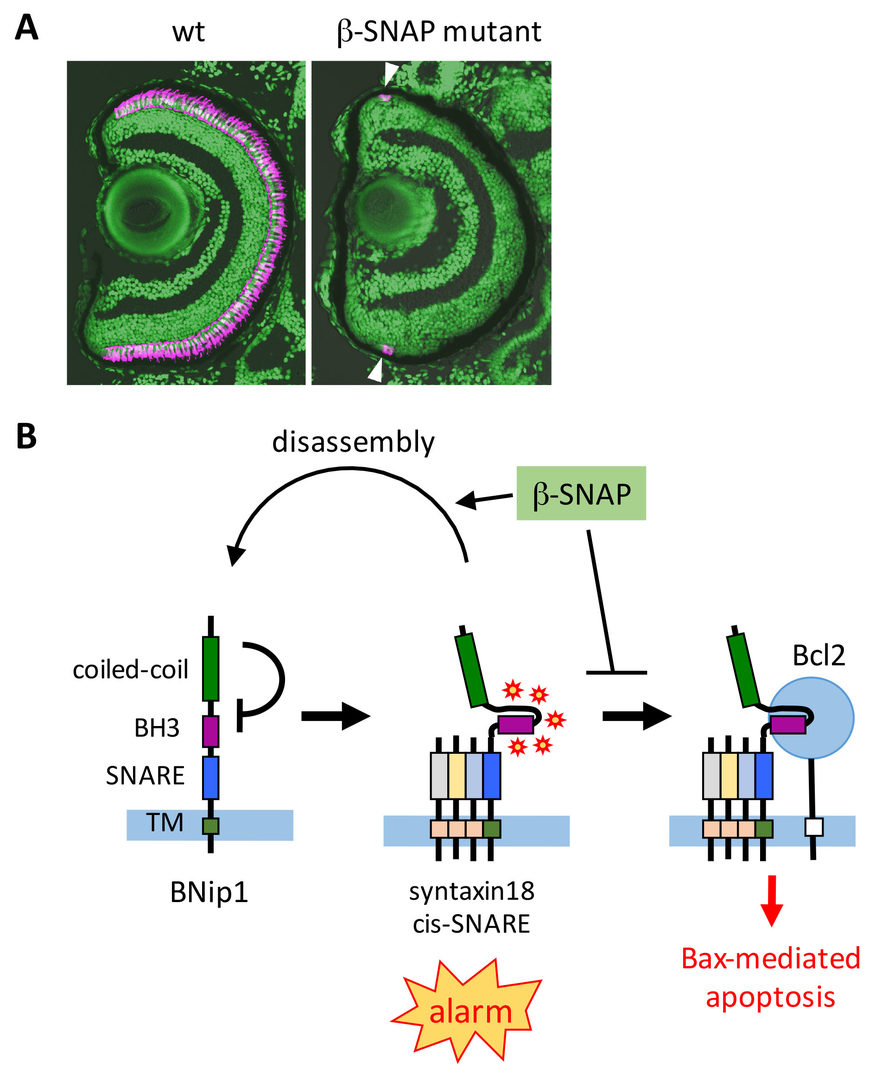
Fig. 1: Surveillance mechanism that monitors vesicular fusion defects in photoreceptor. (A) Wild-type and ß-SNAP mutant retina. Photoreceptors are labeled in magenta. In ß-SNAP mutants, photoreceptors once differentiate (arrowheads) quickly undergo degeneration. (B) Syntaxin-18 cis-SNARE complex functions as an alarm of vesicular fusion defects. ß-SNAP normally disassemble the cis-SNARE complex for SNARE recycle. In b-SNAP mutants, syntaxin-18 cis-SNARE complex is accumulated, and then the BNip1 BH3 domain is activated to induce Bax-dependent apoptosis. This system converts vesicular fusion defects into apoptotic program.
In FY2020, we investigated a physiological role of BNip1-dependent apoptosis. In zebrafish β-SNAP mutants, photoreceptors undergo apoptosis mainly in a small time window of developmental stages, 2 - 4 dpf in which apical photoreceptive membrane structure, called the outer segment (OS) rapidly grows. Since protein synthesis and transport to the OS are highly active in this period, photoreceptor apoptosis in β-SNAP mutants correlates with high levels of protein transport. We examined a critical period of β-SNAP for photoreceptor maintenance by overexpressing b-SNAP under the control of heat shock promoter in β-SNAP mutants (Fig. 2A). We found that overexpression of β-SNAP in 2-5 dpf is enough to prevent photoreceptor apoptosis in zebrafish β-SNAP mutants at least until 21 dpf (Fig. 2B, C).
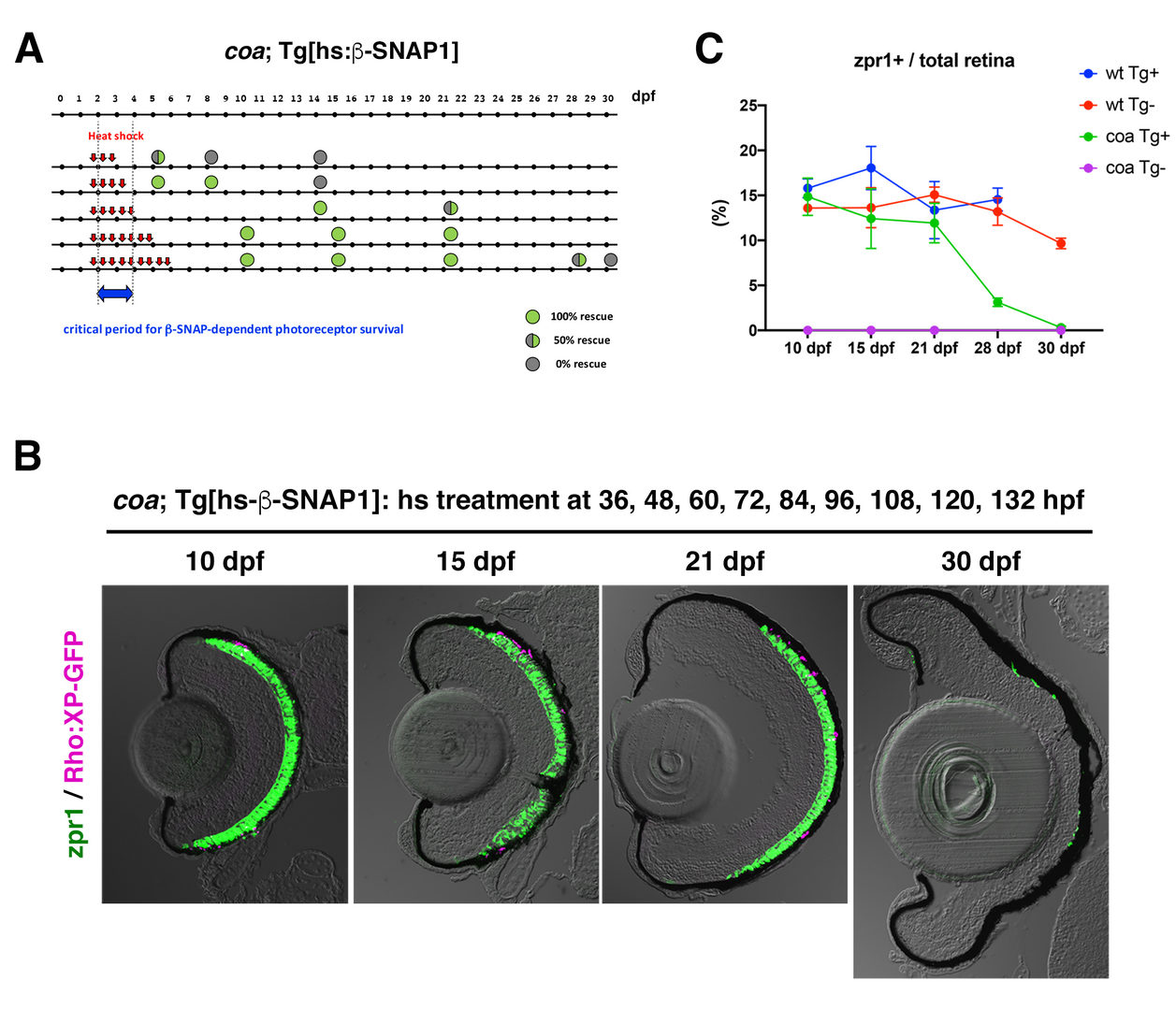
Fig. 2: Transient expression of ß-SNAP1 during the OS growth period is enough to prevent photoreceptor apoptosis in ß-SNAP1 mutants, coa
(A) Experimental design and results of heat shock promoter-driven ß-SNAP1 overexpression in ß-SNAP1 mutants, coa. Zebrafish transgenic line, Tg[hs: ß-SNAP1], were treated with heat treatment at different time points with a 12 hours interval in the period from 36 to 132 hpf. Red arrows indicate one hour-pulsed heat shock treatment at 37oC. Green or grey colored circle indicate the rescue results and its position indicates stage when embryos or larvae were fixed with PFA for analysis. Blue arrow indicates the critical period for ß-SNAP1-dependent photoreceptor survival.
(B) Retinas of coa; Tg[rho:XP:GFP]; Tg[hs: ß-SNAP1] embryos or larvae, which were treated with heat shock nine times at 36, 48, 60, 72, 84, 96, 108, 120, and 132 hpf. Photoreceptors and their OS were visualized by labeling with zpr1 antibody (green) and GFP signals from Tg[rho:XP:GFP] (magenta), respectively. Photoreceptors were maintained at 10, 14, 21 dpf, but degenerated at 30 dpf.
(C) Time line of percentage of zpr1-positive area relative to total retinal area at 10, 15, 21, 28 and 30 dpf. Average percentage of wild-type and coa mutant embryos with and without Tg[hs: ß-SNAP1]. Photoreceptors survived in coa mutant embryos with Tg[hs: ß-SNAP1] by 21 dpf, degenerated at 28 dpf and completely eliminated at 30 dpf (green line).
These data raise the possibility that BNip1-mediated apoptosis links to excessive activation of vesicular transport associated with rapid growth of the OS. Consistently, knockdown of IFT88 and Kif3b, which inhibits transport to the OS, rescued photoreceptor apoptosis in β-SNAP mutants. The treatment of rapamycin, which inhibits protein synthesis through mTOR pathway, also rescued photoreceptor apoptosis in b-SNAP mutants. Importantly, the ER stress response is not activated 2-3 dpf in β-SNAP mutants. Taken together, these data support our model in which BNip1 performs risk assessment that detects excessive vesicular transport in photoreceptors (Fig. 3). In general, deletion of β-SNAP activity compromises recycling of SNARE proteins, which eventually arrests all intracellular protein transport from ER to plasma membrane. In this case, it is likely that the ER stress response is activated to induce apoptosis. However, our study of zebrafish β-SNAP mutants indicates that prior to activation of the ER stress response, BNip1 detects vesicular fusion defects during a rapid OS growth period. Thus, photoreceptors use two different surveillance mechanisms, BNip1 and ER stress response, to monitor the upper and lower ranges of vesicular transport during development.
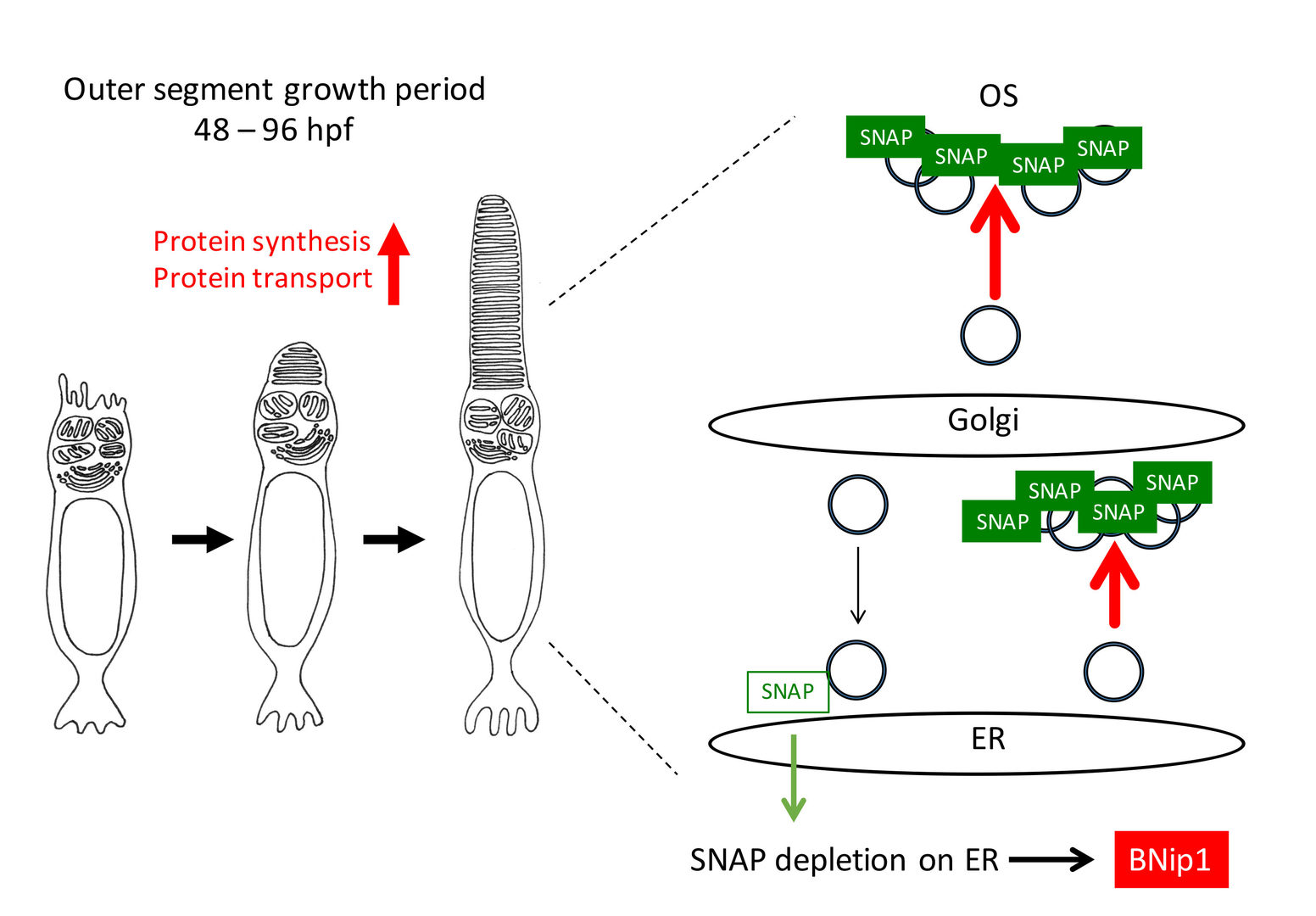
Fig. 3: A model for BNip1-dependent apoptosis. In zebrafish photoreceptors, the OS rapidly grows in 2-4 dpf, suggesting that protein synthesis and transport to the OS are highly activated in this period. Highly activation of vesicular transports may require ß-SNAP molecules in vesicular fusion sites, which subsequently decreases a relative contribution of ß-SNAP on vesicular fusion sites on ER membrane. In this case, it is likely that cis-SNARE complex of syntaxin-18 is accumulated, leading to the activation of BNip1 BH3 domain and Bax activation in mitochondria. In this model, BNip1 monitors excessive activation of vesicular transport during the OS growth period.
3.2 The role of microglia in retinal development and degeneration
Microglia are brain-resident immune cells, originally derived from mesoderm-derivative tissue or the hematopoietic stem cell-lineage, that migrate into brain, and patrol within the brain throughout life. Microglia are thought to eliminate dead or dying neurons to prevent inflammation. Interestingly, microglia-mediated inflammation is required for neuronal regeneration in response to traumatic brain injury in zebrafish (Kyritsis et al., 2012), suggesting more dynamic roles of microglia in neuronal damage. Furthermore, surprisingly, it was reported that microglia facilitate rod cell death in rod-PDE6 mutant mice, which contrasts with our previous view that microglia promote neuronal protection (Zhao et al., 2015). Thus, it is important to understand the role of microglia in photoreceptor degeneration.
We examined normal developmental profiles of microglia and their colonization mechanism to the zebrafish retina. It was reported that microglia are initially generated in lateral plate mesoderm, but that they move to yolk, and enter the brain, including the eyes during zebrafish development (Herbomel et al., 2001). However, it is unknown what kinds of guidance cues enable microglia to move from yolk into developing retina, how microglia are integrated into retinal neural circuits, and also how much cell proliferation contributes to the number of microglia colonizing the retina during development. To answer these questions, we conducted time-lapse observation using zebrafish mpeg1.1:EGFP or mfap4:tdTomato transgenic lines, which specifically visualizes microglia in zebrafish (Ellett et al., 2011; Walton et al., 2015). We found that microglia progressively enter the optic cup from 24 to 54 hpf through the ventral optic fissure. After entry into the optic cup, microglia did not proliferate, suggesting that increased numbers of microglia largely depend on migration from the optic cup. We are now investigating molecular mechanism that guides microglia into optic cup during development.
3.3 Mechanism underlying lens development
The lens is an intraocular organ that focuses visual images on retinal photoreceptors. The lens consists of two cell types: lens epithelial cells and lens fiber cells. Lens epithelium convers the anterior half of the lens fiber core (Lovicu and McAvoy, 2005; Mochizuki and Masai, 2014). At the lens equator, epithelial cells start to differentiate into lens fiber cells, which elongate and pile up to cover the old lens fiber core. Thus, the lens provides a good model for studying spatiotemporal coordination of cell differentiation and morphogenesis (Fig. 4). In vertebrate lens, FGF is secreted from the retina and is believed to form a low-high gradient along the anterior-posterior axis of the lens. It was reported that FGF promotes cell proliferation of lens epithelial cells at low doses and lens fiber differentiation at high doses (McAvoy et al., 2017) (Fig. 4). These observations raised the “FGF gradient hypothesis”, in which FGF regulates multiple steps of lens fiber differentiation: low doses of FGF promote lens epithelial cell proliferation, whereas high doses of FGF induce lens fiber cell differentiation. Furthermore, previous studies revealed a network of transcription factors that coordinate lens epithelial cell proliferation in the anterior lens and lens fiber differentiation in the posterior lens (Fig. 4). These transcription factors are classified into two groups. In the anterior lens epithelium, Pax6 activates Mab21l1 and Foxe3 in concert with Pitx3 and Msx2, resulting in maintenance of E-cadherin expression and a proliferative state of lens epithelial cells. In the posterior lens fiber region, Prox1 activates cell-cycle inhibitor, p57kip2, to promote cell-cycle exit of lens epithelial cells at the equator. In addition, c-Maf and Sox1 activate expression of b- and g-cyrstallin to promote lens fiber maturation. These two groups of transcription factors mutually repress each other to maintain the boundary between the lens epithelium and lens fiber areas (Fig. 4). However, the mechanism that spatially regulates lens epithelial proliferation and lens fiber differentiation is still not fully understood.
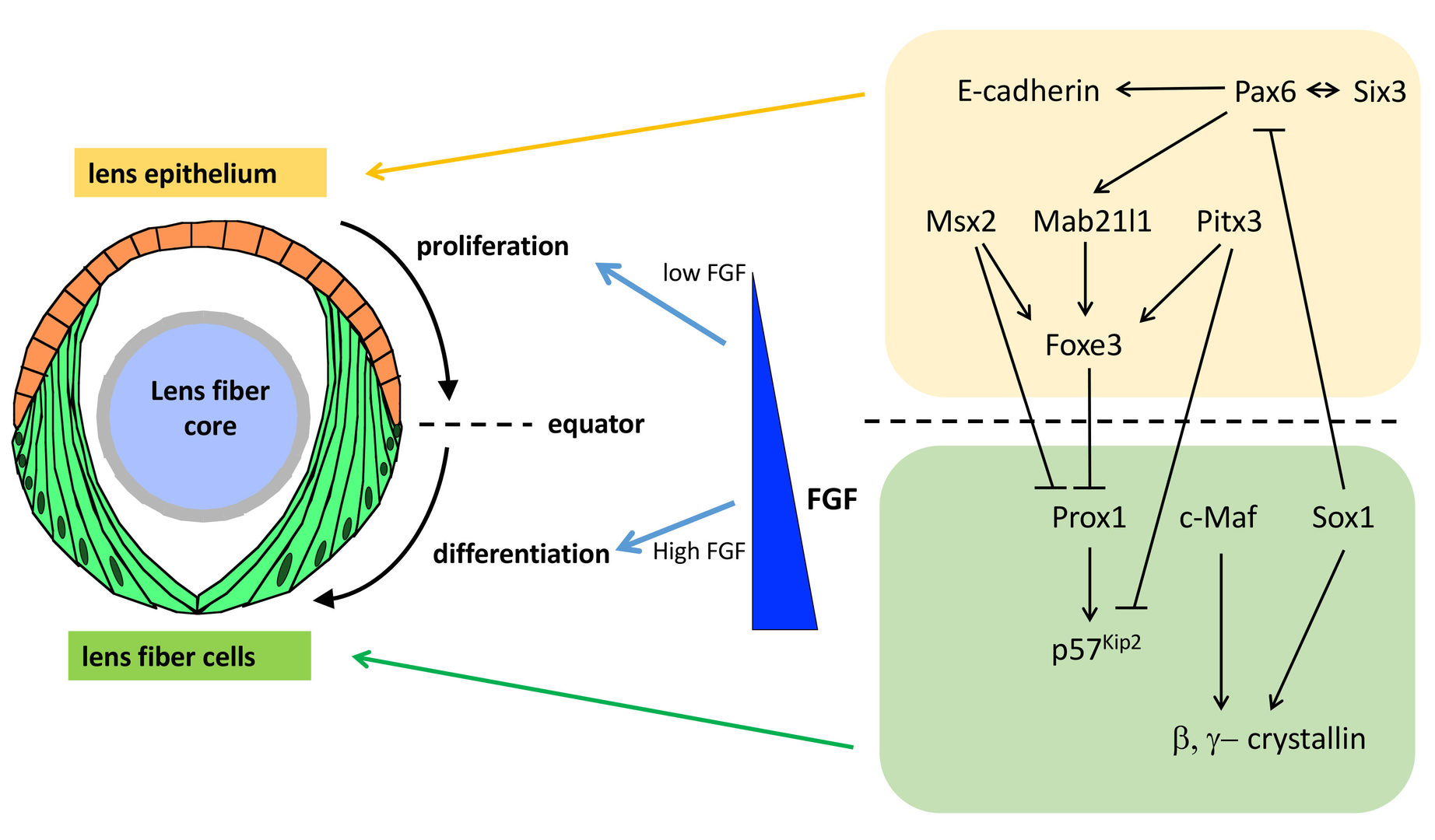
Fig. 4: Mechanism that regulates lens epithelial cell proliferation and lens fiber differentiation. Lens epithelial cells differentiate into lens fiber cells at the lens equator. It was proposed that FGF secreted from the retina forms a low-high gradient along the AP axis of the lens. FGF promotes lens epithelial cell proliferation at low doses, whereas FGF promotes lens fiber differentiation at high doses, supporting the FGF gradient hypothesis. In addition, two groups of transcription factors specify and maintain lens epithelial cell status in the anterior region and lens fiber differentiation status in the posterior region. These two transcription factors mutually suppress each other to keep the equatorial boundary between lens epithelium and lens fiber region.
3.3.1 Mechanism that regulates equator-specific onset of lens fiber differentiation
In order to elucidate the mechanism underlying equator-specific onset of lens fiber differentiation, we focus on one of Fgfrl1 genes, Fgfrl1b, which is specifically expressed in lens epithelium. FGF receptor-like 1 (Fgfrl1) has three extracellular immunoglobulin domains, which bind to FGF ligands, but lacks an intracellular kinase domain. In mammals, proteolytic cleavage of Fgfrl1 releases its ectodomain, which subsequently binds FGF ligands, suggesting that Fgfrl1 functions as a decoy receptor. To inhibit the function of Fgfrl1b, we injected its morpholino antisense into wild-type embryos and examined lens fiber cell differentiation. In Fgfrl1b morphant, expression of an FGF target was elevated, suggesting that Fgfrl1b suppresses FGF signaling. Consistently, the expression of a lens fiber differentiation marker was enhanced. Ectopic differentiation also occurred in lens epithelium of Fgfrl1b morphant. Thus, Fgfrl1b suppresses FGF signaling to ensure equator-specific onset of lens fiber differentiation.
4. Publications
4.1 Journals (* corresponding authors)
- Nishiwaki, Y. and *Masai, I. b-SNAP activity in the outer segment growth period is critical for preventing BNip1-dependent apoptosis in zebrafish photoreceptors. (2020) Sci. Rep. 10 (1): 17379. doi: 10.1038/s41598-020-74360-x.
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
International conference, Oral
None
International conference, Poster
- Babu, S. and Masai, I. Nuclear matrix associated protein, BANP, is required for proper cell-cycle progression and neuronal survival in zebrafish. The 11th European Zebrafish Meeting (Virtual), 2020.
- Kutsia, M and Masai, I. Communication between microglia and adult neural stem cells for neural tissue regeneration in zebrafish model of traumatic brain injury. The 11th European Zebrafish Meeting (Virtual), 2020.
Invited talk
- Sugiyama, Y. and Masai, I. The interaction between basal membrane and cells through mechanical force regulates the formation of lens spherical shape. The 13th RRM (Retina Research Meeting) (Webinar), 12th December 2020.
Domestic conference, Oral
- Nishiwaki, Y., Fujiwara, M. and Masai, I. Dynamin-related GTPase, Drp1, is required for BNip1-mediated photoreceptor apoptosis in zebrafish. JSDB Online Trial Meeting 2020, Japan, 24th – 25th September 2020.
- Babu, S. and Masai, I., Nuclear matrix associated protein, BANP, is required for proper cell-cycle progression and neuronal survival in zebrafish retinas. JSDB Online Trial Meeting 2020, Japan, 24th – 25th September 2020.
- Sugiyama, Y. and Masai, I. Basement membrane and eye lens sculpting. MBSJ2020 online (The 43rd Annual Meeting of the Molecular Biology Society of Japan), 2nd – 4th December 2020.
- Amed, M., Kojima, Y. and Masai, I. Mutation in strip1 gene leads to impaired retinal neural circuit formation in zebrafish, in The 43rd Annual meeting of Japanese Neuroscience Society (Online), 29th July – 1st August 2020.
Domestic conference, Psoter
None
5. Intellectual Property Rights and Other Specific Achievements
5.1 Fundings
HFSP Research Grant
- PI name: Greg Stephens (Vrije Universiteit Amsterdam)
- Co-PI name: Joshua Shaevitz (Princeton Univ)
- Co-PI name: Ichiro Masai (OIST)
- 2016 Sept – 2020 August
KAKENHI (grants from the Ministry of Education, Science and Sport/JSPS)
NA
6. Meetings and Events
6.1 OIST course
- Title: Developmental Neurobiology Course 2020 (eDNC2020)
- Co-organizer: Yoko Yazaki-Sugiyama (OIST), Sam Reiter (OIST), David L. van Vactor (Harvard Medical School, OIST)
- Dates: 23 Feb 2021 – 3 March 2021
- Place: online
- Lecturers: Tom McHugh (RIKEN), Minoru Saitoe (Tokyo Metropolitan Institute of Medical Science), Yoshinori Hayashi (Kyoto Univ.), Todd Roberts (Texas Univ.), Mike Crickmore (Harvard Univ.), Dragana Rogulja (Harvard Univ.), Mineko Kengaku (Koyto Univ.), Aakanksha Singhvi (University Washington School of Medicine, Seattle), Kinichi Nakashima (Kyusyu Univ.), Yulong Li (Chinese Institute for Brain Science, Beijing), Haruhiko Bito (Tokyo Univ.), Marios Chatzigeorgiou (Sar International Centre for Marine Molecular Biology), Gil Levkowit (Weizmann Institute of Science), Maria Tosches (Columbia Univ.), Sang-ki Park (Pohang University of Science and Technolog), Xiaoqun Wang (Chinese Academy Science), C-T Chien (Academia Scinica)
- Tutors: Jelena Katic (OIST), Sarah Morson (OIST), Makoto Araki (OIST), Yuichi Morohashi (OIST), Luis Carretero (OIST)
6.2 OIST Workshop
None
6.3 OIST Seminars
None
7. Other
Nothing to report.



