Cell Proliferation and Gene Editing Unit (Franz Meitinger)
Research Interests
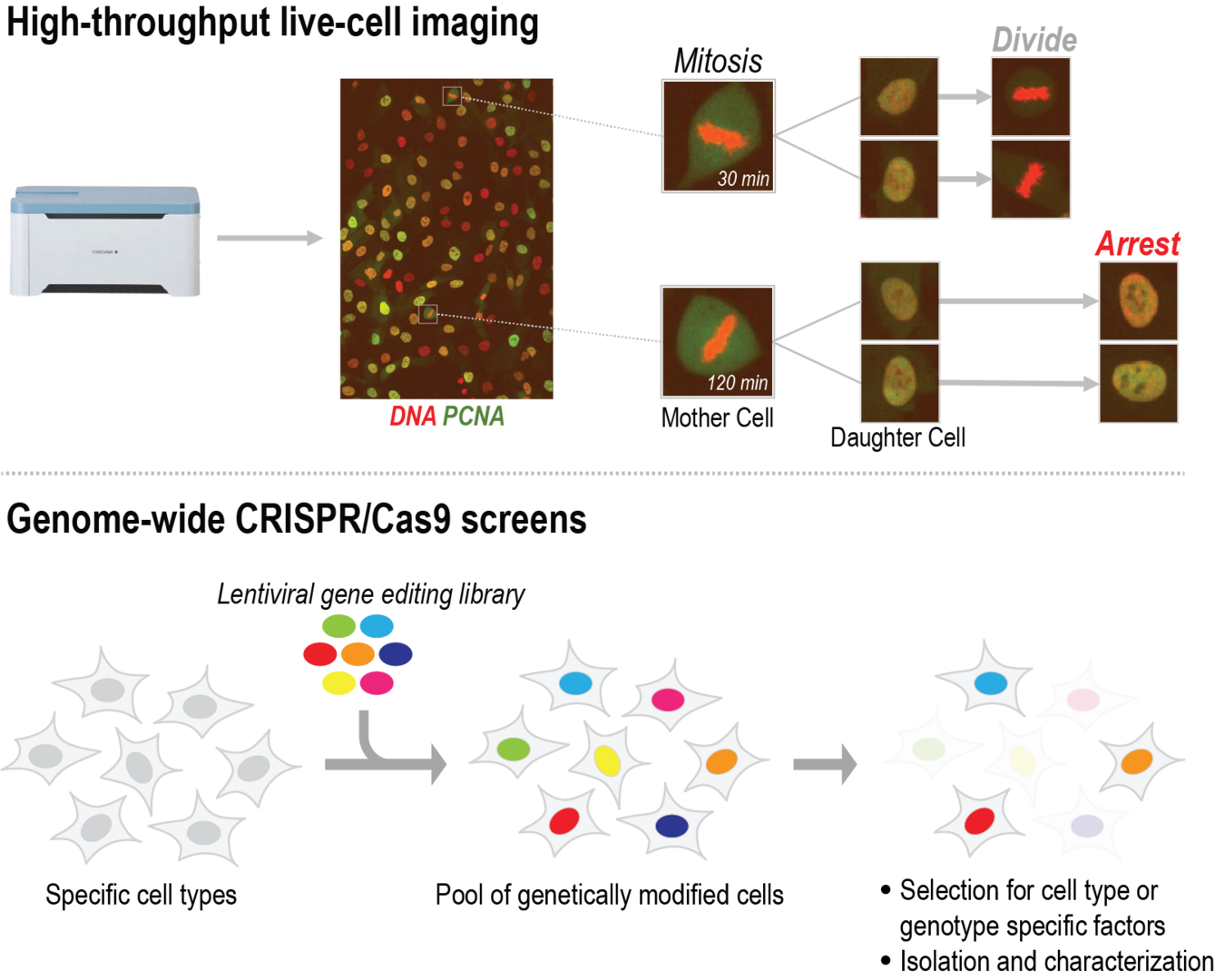
Every day, millions of cells in our body divide to maintain essential tissue functions. Errors in cell division can lead to developmental disorders or cancer. Our research focuses on molecular mechanisms of cell division and quality control in normal and cancer cells in order to understand cell type-specific and tumor-suppressive mechanisms and to identify biomarkers that confer a cancer-specific vulnerability to chemical drugs. We combine high throughput imaging, gene editing, genome-wide screens, and biochemistry to open new avenues for therapeutic development.
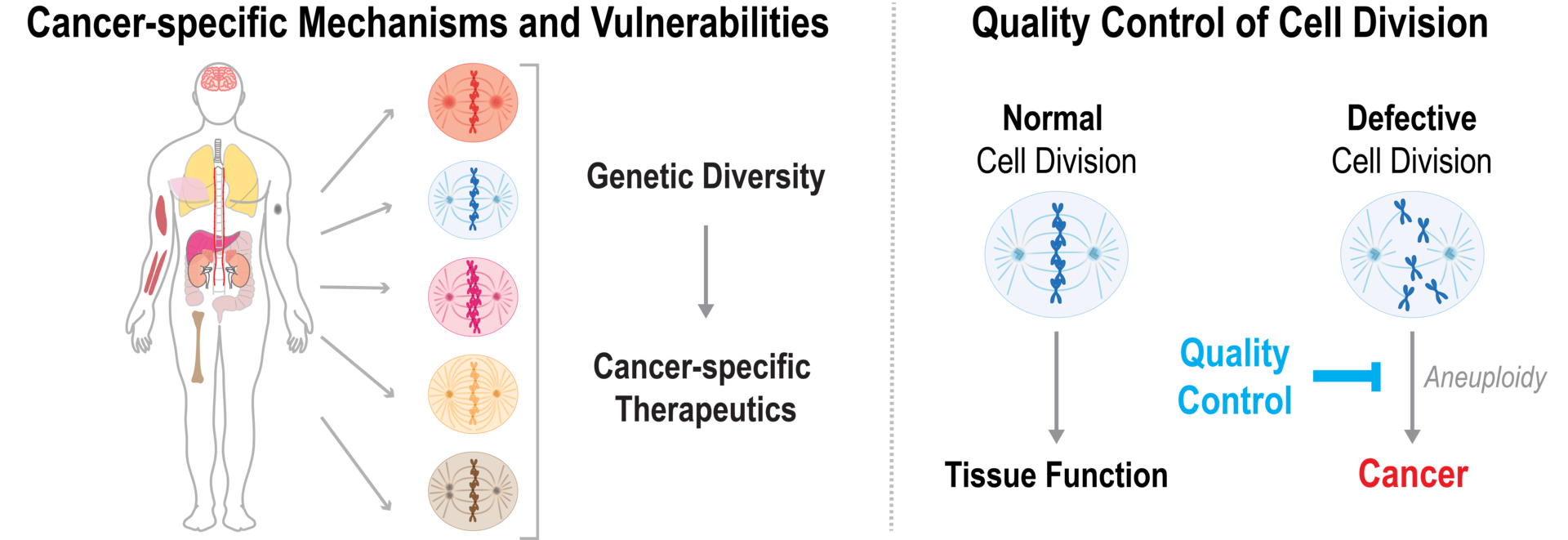
1. Cancer-specific Mechanisms and Vulnerabilities
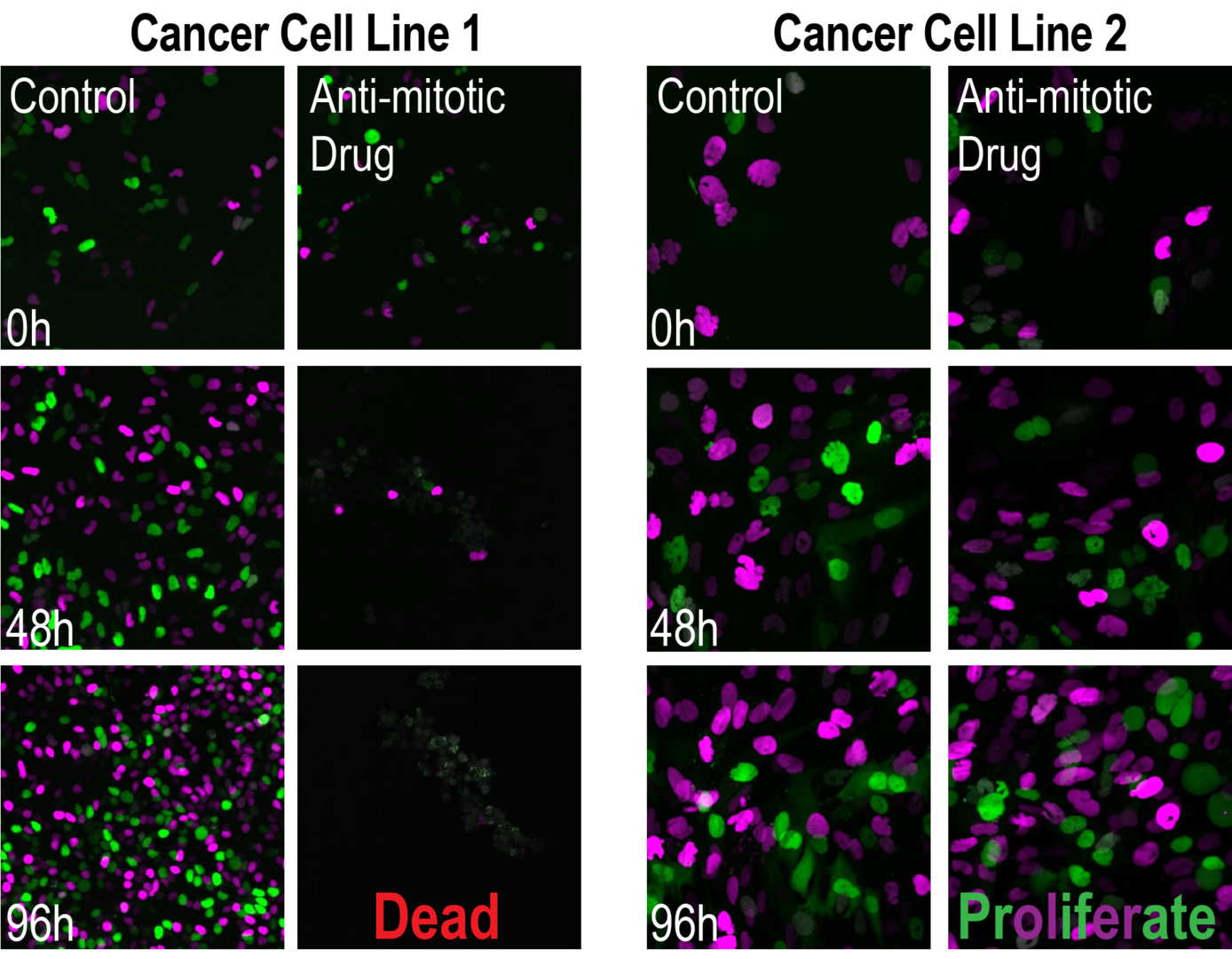
Most approved chemotherapies also target healthy cells, which can lead to severe health problems. Therefore, it is desirable to develop therapeutic approaches that specifically target cancer cells. Many cancer cells acquire cancer-specific mutations or chromosomal alterations that allow them to continue to reproduce without any restriction. These alterations confer specific properties that may accompany vulnerabilities that can be targeted in therapeutic approaches. Our goal is to identify cancer-specific vulnerabilities that open new opportunities for therapeutic development (Meitinger et al. 2020, Nature; Meitinger et al. 2024, Science)
- Meitinger, F.#, Belal, H., Davis, R.L., Martinez, M.B., Shiau, A.K., Oegema, K.#, Desai, A.# (2024). Control of cell proliferation by memories of mitosis. Science 383 (6690):1441-1448. # corresponding authors
- Meitinger, F.#, Ohta, M., Lee, K.Y., Watanabe, S., Davis, R.L., Anzola, J.V., Kabeche, R., Jenkins, J., Shiau, A.K., Desai, A.#, Oegema, K.# (2020). The ubiquitin ligase TRIM37 controls cancer-specific vulnerability to PLK4 inhibition. Nature 585:440-446. # corresponding authors
2. Quality Control of Cell Division to Maintain Genome Stability
Defects in cell division can lead to aneuploidy and tumor development. To prevent the catastrophic consequences of cell division defects, proliferating cells continuously examine the integrity of their organelles, molecular machines of cell division, and the genetic information saved on the chromosomes. Tumor suppressors like p53 recognize defects and stop the proliferation of harmed and potentially dangerous cells.
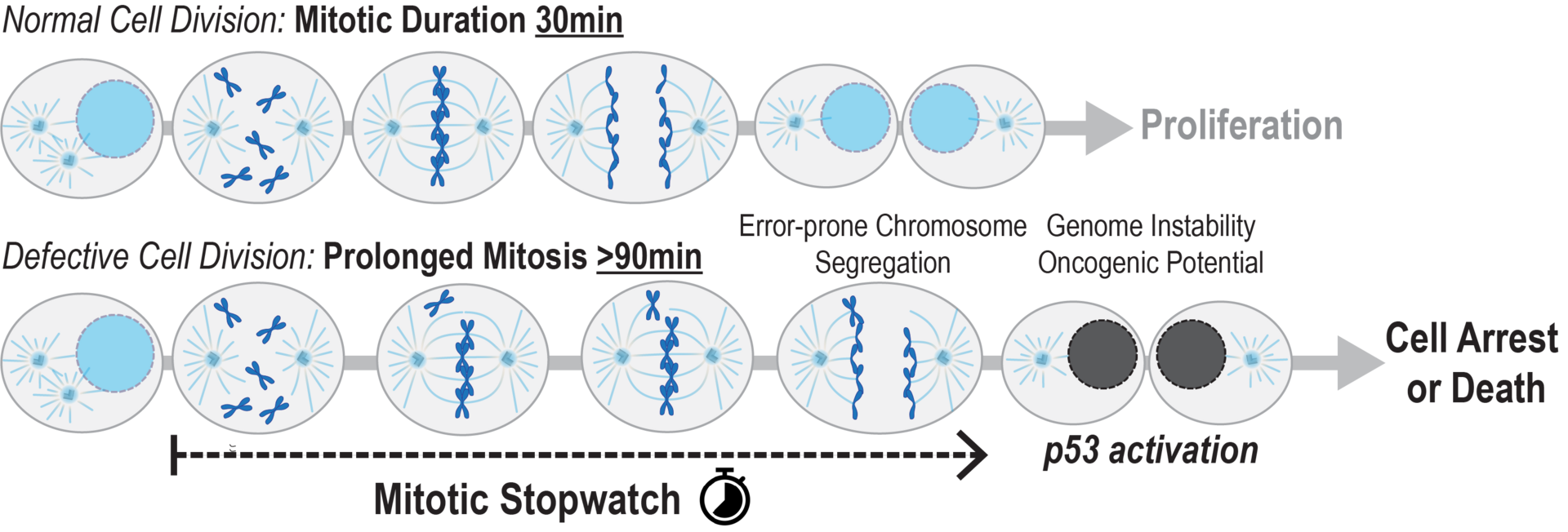
Mitosis is one of the most critical cell cycle phases. During mitosis, the duplicated DNA condenses into compact chromosomes that segregate to the two daughter cells. This process is controlled by the mitotic machinery that consists of a microtubule-based spindle and centrosomes, which are the major microtubule-organizing centers. We are interested in mechanisms that faithfully segregate chromosomes and protect from defects that can lead to aneuploidy and cancer development.
We recently discovered a mitotic surveillance mechanism—the Mitotic Stopwatch—that detects defects in cell division by measuring the duration of mitosis (Meitinger et al. 2016 JCB, Meitinger et al. 2024 Science). An extended mitotic duration indicates that cells have problems in faithfully distributing their genetic information to the daughter cells. Our work suggests that the Mitotic Stopwatch detects potentially dangerous cells, which are prone to accumulate aneuploidy, and initiates a p53-dependent cell arrest or death. Our aim is to understand the molecular mechanism of the Mitotic Stopwatch and how it protects the organism from cancer development.
- Meitinger, F.#, Belal, H., Davis, R.L., Martinez, M.B., Shiau, A.K., Oegema, K.#, Desai, A.# (2024). Control of cell proliferation by memories of mitosis. Science 383 (6690):1441-1448. # corresponding authors
- Meitinger, F., Anzola, J.V., Kaulich, M., Richardson, A., Stender, J.D., Benner, C., Glass, C.K., Dowdy, S.F., Desai, A., Shiau, A.K., Oegema, K. (2016). 53BP1 and USP28 mediate p53 activation and G1 arrest after centrosome loss or extended mitotic duration. Journal of Cell Biology 214, 155-166.
Methods
Our unit combines high throughput methods such as genome-wide screens as well as gene editing, live-cell imaging, and biochemistry to identify novel cancer-specific vulnerabilities using untransformed and a variety of childhood and adult cancer cell lines as model systems.