FY2021 Annual Report
Marine Eco-Evo-Devo Unit
Professor Vincent Laudet

(From left to right)Yoko Fujitomi,,James Hutasoit,Marleen Klann,Vincent Laudet,Marcela Herrera Sarrias,Jann Zwahlen,Manon Mercader,Hiroki Takamiyagi,Ken Maeda,Makiko Ajimura,Saori Miura,Emma Gairin,Kina Hayashi, Rio Kashimoto
Abstract
The Marine Eco-Evo-Devo Unit uses the extraordinary diversity of coral reef fish to investigate the evolution of life history strategies in marine fishes, integrating ecological, evolutionary and developmental components. Using anemonefish as model systems we study the function, development and evolution of color patterns and how environment acting on hormonal pathways change those patterns.
The year 2021 was only the second year of the MEEDU unit! Because of the pandemic we still had no complete husbandry, strongly limiting the developmental experiments we could carried out. The work developed very well on the genomic side as well as on molecular ecology with a very nice collaboration with Tim Ravasi unit. We are now 17 people in the unit and there is quite a diversity of topics covered.
The unit has also started in our second location at Academia Sinica in Taiwan at Yilan Marine Research Station of ICOB (Institute of Cellular and Organismic Biology). Thanks to the work of Fiona Lee and Kai Wu there we have made tremendous progress on functional analysis of mutant anemonefish
1. Staff
- Dr. Marleen Klann, Staff Scientist
- Dr Ken Maeda, Staff Scientist
- Dr. Manon Mercader, Postdoctoral Scholar
- Dr. Marcela Herrera Sarrias, Postdoctoral Scholar
- Dr. Saori Miura, Lab Manager
- Dr. Kina Hayashi, Research Fellow
- Lilian Carlu, Husbandry Technician
- Hiroki Takamiyagi, Fieldwork Technician
- James Hutasoit, Research Assistant
- Mathieu Reynaud, Special Research Student
- Emma Gairin, PhD Student
- Jann Zwahlen, PhD Student
- Yuki Tara, PhD Student
- Rio Kashimoto, PhD Student
- Agneesh Barua, PhD Student
- Yoko Fujitomi, Research Unit Administrator
- Makiko Ajimura, Research Unit Administrator
2. Collaborations
2.1 Sea anemone genomics
- Mutualism between giant sea anemone and anemonefish ancestors is believed to have started 10 to 12 million years ago. Even though the interest of this symbiosis is immediately clear in terms of protection for the anemonefish partners, much less is known about the advantage and constrains of this association for the sea anemone. To understand the mechanisms and evolutionary strategy of this symbiotic relationship for the sea anemone in collaboration with Satoh unit we decided to bring new information on giant sea anemones. Rio Kashimoto with Konstantin Khalturin in Satoh unit performed a transcriptomic analysis of the 7 species of giant sea anemones living in Okinawa to gain insight on the genome composition and gene content of these essential anemonefish partners. This analysis has now been published in Zoological Science and we are currently determining the complete genomes of two different types of giant sea anemone. Furthermore, in collaboration with James Reimer at the university of the Ryukyus, we are using a phylotranscriptomic analysis to better understand the diversity of giant sea anemones. Lastly, we study the effects that anemone fish have on the giant sea anemones.
- Type of collaboration: Joint research
- Researchers:
- Professor Nori Satoh, OIST
- Dr Konstantin Khalturin, Satoh unit
- Rio Kashimoto, PhD student
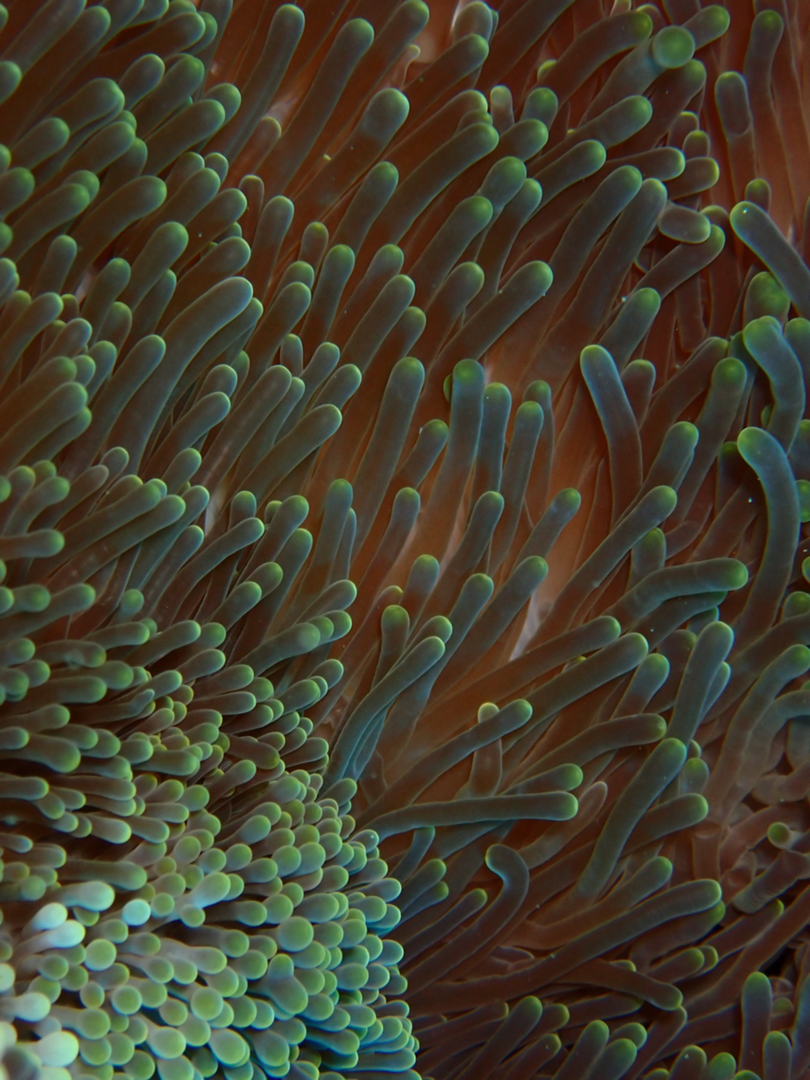
A giant sea anemone, Stichodactyla mertensii
2.2 Ecological types of anemonefish
- Understanding the reason why there is such a wide diversity of species is a major goal in evolution. But it is often quite difficult to understand the precise ecological differences existing between related species. In collaboration with Marco Rosti unit we used the six anemonefish species living in Okinawa as models, and by a combination of various approaches we investigated their swimming abilities to see if we could define distinct ecological types. Manon Mercader first measures fish critical swimming speed and metabolic rate. Then micro-CT scan of the different species were determined in collaboration with OIST imaging section and from these images their 3D shape can be derived. Finally, water flow around the fish was modelized by Stefano Olivieri in Marco Rosti unit to get insight in how the shape might affect how the fish swims and the associated cost of transport. From this we concluded that we can define several ecological types that on which we can allocate each of the 28 species of anemonefish existing in the indo-pacific area..
- Type of collaboration: Joint research
- Researchers:
- Professor Marco Rosti, OIST
- Dr. Manon Mercader, post-doc, Laudet unit
- Dr. Stefano Olivieri, post-doc, Rosti unit
- Shinya Komoto OIST imaging section microCT Scan
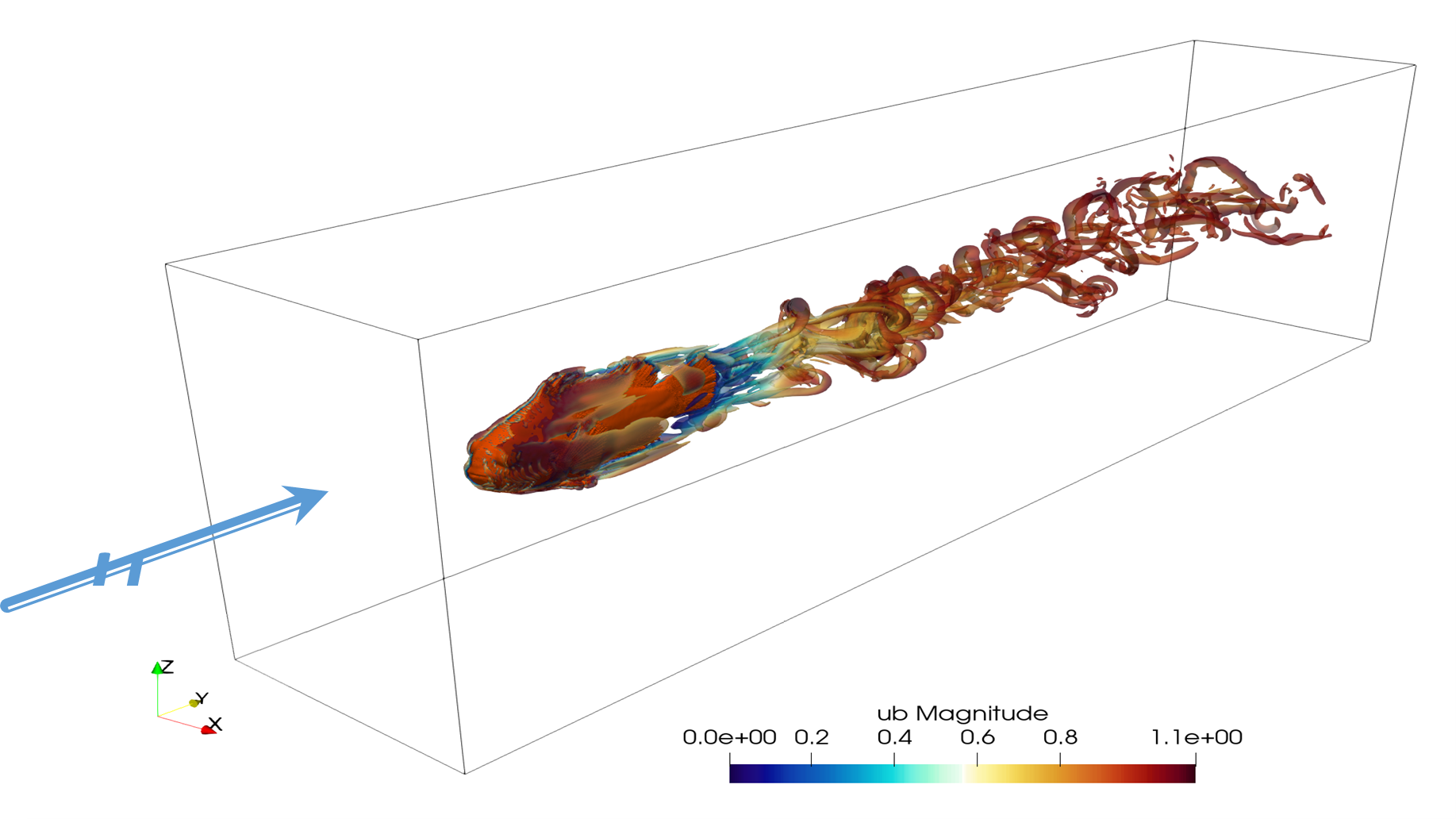
The turbulence generated by the fish shape on a flow of water allow to measure the drag force and to compare it between species.
3. Activities and Findings
3.1 Chromosome-scale genome assemblies of two anemonefish species from Okinawa
Anemonefish are an emerging model organism for studying the ecology, evolution, adaptation, and developmental biology of reef fishes. Yet, high-quality genomic resources are still scarce, hindering advanced genomic analyses. Leveraging the power of PacBio long-read sequencing and Hi-C chromosome conformation capture techniques, we constructed a high-quality chromosome-scale genome assembly for the clownfish Amphiprion ocellaris, the main character of the Disney movie “Finding Nemo”. Together with its sister species Amphiprion percula, these two species are separated from the other 26 anemonefish. Our results revealed a high number of genes conserved only in the two clownfish and not in other species. Interestingly, we found that most of these genes are related to neurological functions and exhibited distinct expression patterns in brain tissue, highlighting the genetic toolkits involved in the unique ecology and behavior of the two clownfish compared to the rest of anemonefish. Similarly, we also assembled the genome of the yellowtail anemonefish Amphiprion clarkii, a species known for being highly polymorphic in their melanistic coloration (black pigmentation). Detailed analysis of pigmentation genes showed that this species has two additional copies of a tyrosine-protein kinase, a gene involved in melanophore development and pigment pattern formation. Overall, our studies provide the highest quality genomes for anemonefish to date whilst also providing a valuable resource for understanding the ecology and evolution of reef fishes.
Type of collaboration: Joint research
Researchers:
- Vincent Laudet, Marine Eco-Evo-Devo Unit, OIST
- Timothy Ravasi, Marine Climate Change Unit, OIST
- Taewoo Ryu, Research Scientist, Marine Climate Change Unit
- Marcela Herrera Sarrias, post-doc, Marine Eco-Evo-Devo Unit
- Billy Moore, PhD student, Marine Climate Change Unit
Publication:
Ryu T*, Herrera M*, Moore B, Izumiyama M, Kawai E, Laudet V, and Ravasi T. 2022. A chromosome-scale genome assembly of the false clownfish, Amphiprion ocellaris. G3 Genes|Genomes|Genetics: 10.1093/g3journal/jkac074
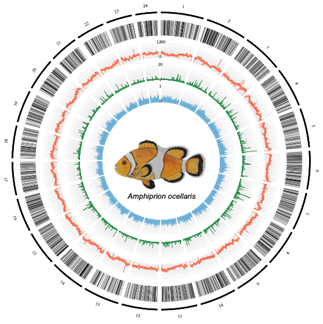
Chromosome architecture of the Amphiprion ocellaris genome. From the outermost layer inwards, each layer represents (1) chromosomes indicated by lines and ordered by size; (2) genic regions; (3) the number of repeats per 100 kb; (4) the PhyloP score calculated from the whole-genome alignment of anemonefishes with Acanthochromis polyacanthus as an outgroup species; and (5) tissue-specificity index (τ) of each gene
3.2 Color patterns of anemonefish
Color pattern analysis in anemonefish, displaying 1-3 white bars with a black edge on an orange-reddish body, is promising and has already provided valuable insights into several areas of pigmentation research. We previously discovered that 3 pigment cell types contribute to anemonefish coloring: black melanophores, yellow/orange xanthophores, and white iridophores. To explore the relationship between those three cell types we employ pharmacological treatment to alter differing aspects of individual cell types. For example, if fish are treated with DPAT, a compound that inhibits melanophore migration, we find that white bar formation is disrupted resulting in incompletely formed bars (B row in figure below). Additionally, we use color mutants to gain more information, like “Darwin Black” (C row in figure below) which as adults are black instead of orange. Interestingly, during color development their bars are forming incompletely at first, similarly to DPAT treatments. Comparing both data sets suggests that a high density of black melanophores restricts the movement of white iridophores, preventing normal white bar establishment.

Increased melanophore density disrupts white bar formation. (A1-A3) White bar formation in control fish. The white bar develops as a continues bar. (B1-B3) When melanophore migration is inhibited (DPAT treatment) the white bar is forming dis-continuously. (C1-C3) In “Darwin Black” mutants, that have a higher number of melanophores, the white bar is also forming incompletely, from the dorsal and ventral part, eventually meeting in the middle.
3.3 Stripes and bars in anemonefish colonies
The spectacular pigment pattern of coral reef fish has since a long time attracted the attention of biologists and this have led to a substantial number of possible explanations. Among coral reef fishes, anemonefish recently became a model to study pigmentation and two major hypotheses are currently discussed for the function of these brilliant colors and in particular the conspicuous white bars they harbor. The first hypothesis is that these colors are used as a system to warn predators of the toxicity of the sea anemone hosting the fish. Whereas the second assumes that white bars are used as a system for species recognition and more generally have social functions in the anemonefish colonies. In our paper, by exploiting the rich fish fauna associated with sea anemones we provide evidence in favor of this second hypothesis. Indeed, working in the Ryukyu Archipelago we bring two complementary evidences: (i) By comparing the fish biota associated with either sea anemone (namely Stichodactyla gigantea inhabited by Amphiprion ocellaris) or scleractinian corals and their color pattern we reveal that around host anemones and zooxanthellate there was no fish species with bars but the fish species with stripe is more abundant suggesting the possibility that anemonefish actively repels as competitors fish harboring vertical bars. (ii) we directly test experimentally this model in the field by showing that the dominant A. ocellaris females responded more aggressively to intruders with bar patterns than those with stripe patterns, hence protecting their host anemone as their own territory. This indicates that only fish species without bar patterns may have access to host anemones (Fig. 1).
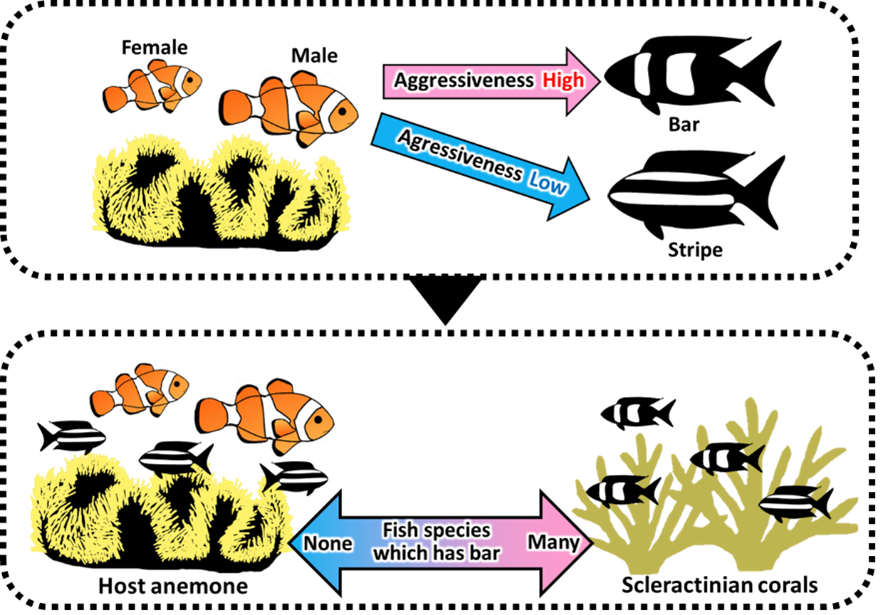
Schematic drawing of the relationship between differences in aggressive behavior of anemonefish toward bar and stripe patterns and differences in color patterns of fish inhabit coral and host anemone.
3.4 Do color-morphs of goby represent different species?
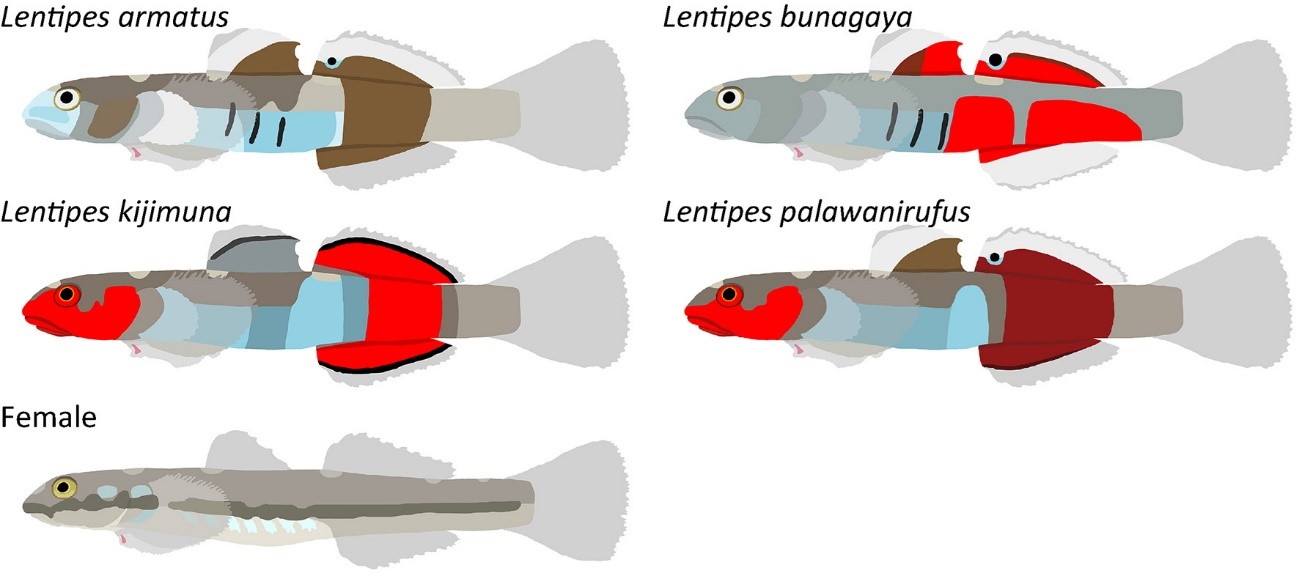
Body color is one of the major characters to distinguish fish species. It is often used as an essential criterion to identify fishes at the species level in many taxa. On the other hand, distinctive color morphs within a single species are often known. We studied morphology and genetics of Lentipes gobies collected in Japan and the Philippines. Despite the fact that males were classified into four color-morphs, no morphological differences other than the characteristic color pattern could be found. These four morphs were also not distinguished by mitochondrial genome sequences. Genomewide SNPs analysis clearly separated them, suggesting that they indeed represent four independent lineages. We observed that males display their species-specific body colorations during courtship. Prezygotic isolation due to female preferences for different male body colors is probably the primary mechanism of reproductive isolation between the four species. Three of the four species were described as new species and two of them, Lentipes kijimuna and L. bunagaya were featured in “New Species 2021” report.
https://shoalconservation.org/new-species-2021-report-released/
We also published two new species of Rhinogobius from Palawan, Philippines. They were featured in following Mongabay news.
4. Publications
4.1 Journals
- Maeda K, Kobayashi H, Palla HP, Shinzato C, Koyanagi R, Montenegro J, Nagano, Atsushi J, Saeki T, Kunishima T, Mukai T, Tachihara K, Laudet V, Satoh N, Yamahira, K. Do colour-morphs of an amphidromous goby represent different species? Taxonomy of Lentipes (Gobiiformes) from Japan and Palawan, Philippines, with phylogenomic approaches. Systematics and Biodiversity 2021, 19: 1080-1112.
- Salis P, Lee, S-H, Roux N, Lecchini D, Laudet V. The real Nemo movie: filmed embryonic development of Amphiprion ocellaris from first division to hatching. Developmental Dynamics, 2021, 250: 1651-1667.
- Salis P, Roux N, Huang D, Marcionetti A, Mouginot P, Reynaud M, Salles O, Salamin N, Pujol B, Parichy DM, Planes S, Laudet V. Thyroid hormones control the formation and plasticity of white bars in clownfishes. Proc. Natl. Acad. Sci. USA, 2021, 118 : e2101634118.
- Beinsteiner B, Markov GV, Erb S, Chebaro Y, McEwen A, Cianférani S, Laudet V, Moras D and Billas IML. A structural signature motif enlightens the origin and diversification of Nuclear Receptors. Plos Genetics. 2021, 4, e1009492.
- Roux N, Logeux V, Trouillard N, Pillot R, Magré K, Salis P, Lecchini D, Besseau L, Laudet V, Romans P. A star is born again: Methods for larval rearing of an emerging model organism, the clownfish Amphiprion ocellaris. Journal of Experimental Zoology B, 2021 336: 376-385.
- Klann, M, Mercader M, Carlu L, Hayashi K, Reimer J, Laudet V. Variation on a theme: Pigmentation variants and mutants of anemonefish. EvoDevo, 2021 12: 8.
- Jessus C, Laudet V. Edouard Chatton, arpenteur des mondes minuscules. Pour la Science, 2022, 532: 74-81.
- Markov GV, Laudet V. Small molecules as products of evolution. Current Biology, 2022, 32, R97–R115.
- Yamahira K, Ansai S, Kakioka R, Yaguchi H, Kon T, Montenegro J, Kobayashi H, Fujimoto S, Kimura R, Takehana Y, Setiamarga DHE, Takami Y, Tanaka R, Maeda K, Tran HD, Koizumi N, Morioka S, Bounsong V, Watanabe K, Musikasinthorn P, Tun S, Yun LKC, Masengi KWA, Anoop VK, Raghavan R, Kitano J. Mesozoic origin and ‘out-of-India’ radiation of ricefishes (Adrianichthyidae). Biology Letters 17: 20210212 (2021).
- Kunishima T, Maeda K, Inui R, Hibino Y. First Japanese record of Muraenichthys gymnopterus (Anguilliformes, Ophichthidae) from Ishigaki-jima Island, Ryukyu Archipelago. Species Diversity 26(2): 343–349 (2021).
- Maeda K, Shinzato C, Koyanagi R, Kunishima T, Kobayashi H, Satoh N, Palla HP. Two new species of Rhinogobius (Gobiiformes: Oxudercidae) from Palawan, Philippines, with their phylogenetic placement. Zootaxa 5068(1): 81–98 (2021).
- Kunishima T, Palla HP, Tachihara K, Maeda K. First Records of an Estuarine Goby Acentrogobius ocyurus (Gobiiformes: Gobiidae) from Japan and the Sulu Sea in the Philippines. Species Diversity 27: 129–138 (2022).
4.2 Books and other one-time publications
Klann M, Mercader M, Salis P, Reynaud M, Roux N, Laudet V, Besseau L. Anemonefishes. In Established and Emerging Marine organisms in experimental biology edited by B. Schierwater and A. Boutet, CRC Press. 443-463
4.3 Oral and Poster Presentations
- Laudet V. « Coral Reef Fishes as models for Eco-Evo-Devo» Assemble+ general assembly virtual meeting January 28th, 2021
- Laudet V. “Fish models: From zebrafish to clownfish as an inspiration to increase knowledge in cosmetics” My Blue Cosmetic. Banyuls/Mer, France (virtual presentation) March 10-12th, 2021
- Maeda K. Revision of Eleotris species described by Bleeker. Meetiong of Fish Taxonomy Society. Online, 20 December, 2021. (Oral)
- Iida M, Kobayashi H, Tran HD, Maeda K. Diadromous migration of gobies in Vietnam in comparison with other Asian regions. The 2nd National Conference on Ichthyology in Vietnam. Online, 25 December 2021. (Oral)
- Maeda K, Kobayashi H, Yamakawa U, Shinzato C, Palla HP, Tran HD. Taxonomic revision of the genus Eleotris in the western Pacific. The 55th Annual Meeting of the Ichthyological Society of Japan. Online, 18 September 2021. (Oral)
- Kunishima T, Tachihara K, Palla HP, Maeda K. First Japanese record of a goby species, Drombus ocyurus, collected from Manko, Okinawajima Island. The 55th Annual Meeting of the Ichthyological Society of Japan. Online, 18–19 September 2021. (Poster)
- Kobayashi H, Yamakawa U, Sato M, Maeda K, Yamahira K. Phylogeny and taxonomic revision of the Mangrove Giant Gudgeon, genus Ophiocara in the Ryukyu Archipelago, Japan. The 58th Annual Meeting of the Biological Society of Okinawa. Online, 29 May 2021. (Oral)
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
Nothing to report.
7. Other
Nothing to report.



