FY2020
Plant Epigenetics Unit
Associate Professor Hidetoshi Saze

Abstract
Epigenetic Regulation of Genes and Transposable Elements in Plant Genomes
We are studying epigenetic regulation of genes and transposable elements (TEs) in plant genomes. Genome defense mechanisms in plants repress TEs by epigenetic modifications, such as DNA cytosine methylation, small RNAs, and modifications of histone proteins. In contrast, these repressive epigenetic modifications are generally excluded from actively transcribed genes. The major goal of our research is to understand how epigenetic mechanisms distinguish gene and TE sequences, how they deposit specific chromatin modifications at the targets, and to elucidate their biological significance in environmental adaptation and genome evolution.
1. Staff
- Hidetoshi Saze, Associate Professor
- Matin Miryeganeh, Researcher (JSPS PD)
- Leonardo Furci, Researcher
- Jeremy Berthelier, Researcher
- Oscar Juez Neira, Student
- Munissa Sadykova, Student
- Yoshiko Harukawa, Technical Staff
- Tomoe Shimazaki, Technical Staff
- Yoko Fujitomi, Research Administrator
2. Collaborations
2.1 Genome analysis of Fukugi tree in Okinawa
- Type of collaboration: Joint research
- Researchers: Ms. Ayaka Irei, Okinawa Prefectural Agricultural Research Center
2.2 Transposable elements in plant genomes
- Type of collaboration: Joint research
- Researchers: Dr. Akira Kawabe, Faculty of Life Sciences, Kyoto Sangyo University
2.3 Functional Okinawa Rice Project
- Type of collaboration: Joint research
- Researchers: Onna Village
3. Activities and Findings
3.1 Epigenetic regulation of spurious transcription initiation in Arabidopsis
In plants, epigenetic regulation is critical for silencing transposons and maintaining proper gene expression. However, its impact on the genome-wide transcription initiation landscape remains elusive. By conducting a genome-wide analysis of transcription start sites (TSSs) using cap analysis of gene expression (CAGE) sequencing, we show that thousands of TSSs are exclusively activated in various epigenetic mutants of Arabidopsis thaliana. Many have not been identified in previous studies, of which up to 65% are contributed by transposons. They possess similar genetic features to regular TSSs and their activation is strongly associated with the ectopic recruitment of RNAPII machinery. The activation of cryptic TSSs significantly alters transcription of nearby TSSs, including those of genes important for development and stress responses. Our study, therefore, sheds light on the role of epigenetic regulation in maintaining proper gene functions in plants by suppressing transcription from cryptic TSSs.
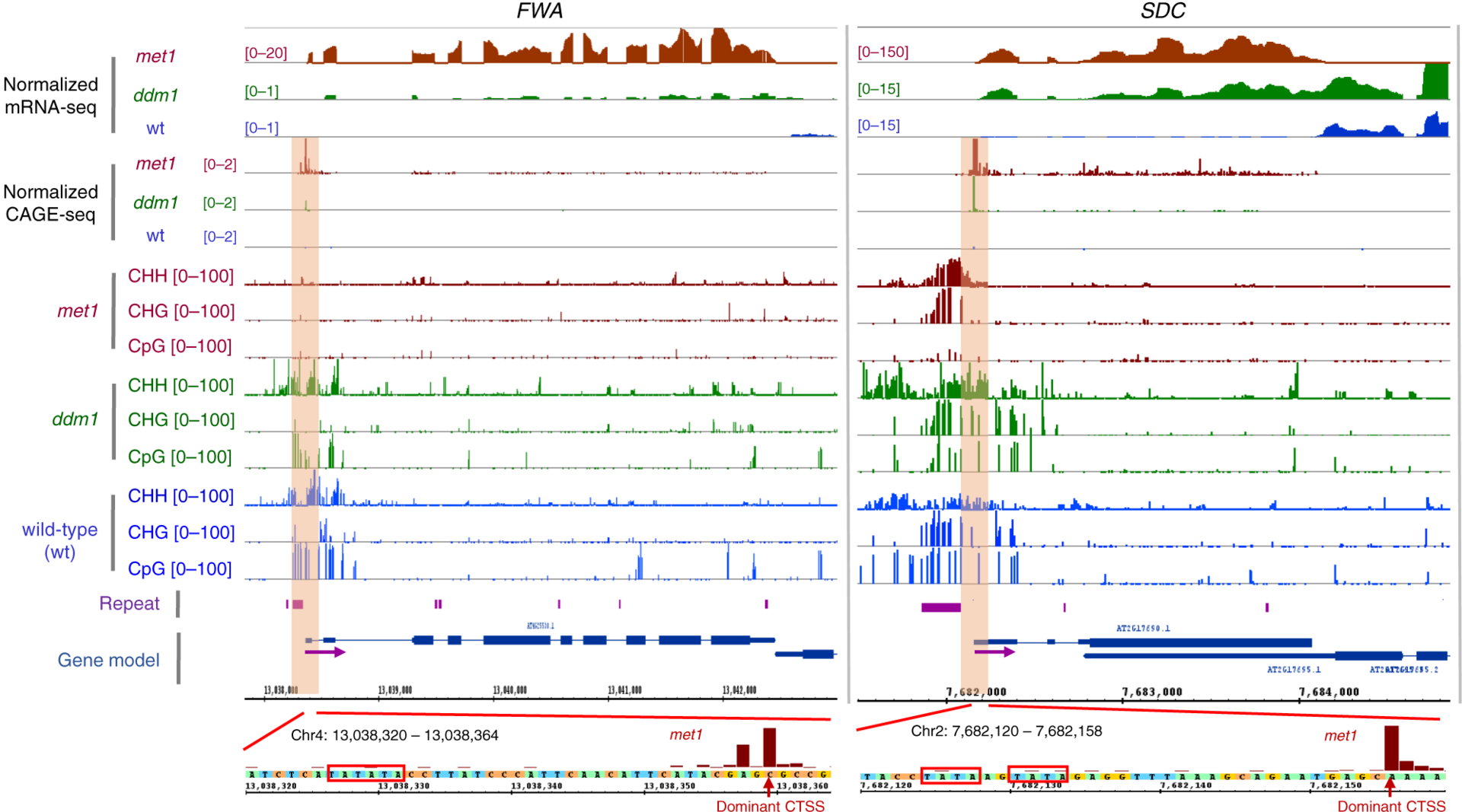
Figure1: Browser tracks showing the ectopic activation of the TSSs at FWA (AT4G25530) and SDC (AT2G17690) gene loci in the met1 and ddm1 backgrounds (indicated by orange windows). Their dominant CTSSs (red arrows) were aligned to the upstream sequences, indicating the presence of TATA-box motifs (red boxes) encoded within (left panel) or downstream (right panel) of the nearby repeats. Purple arrows indicate the direction of transcription.
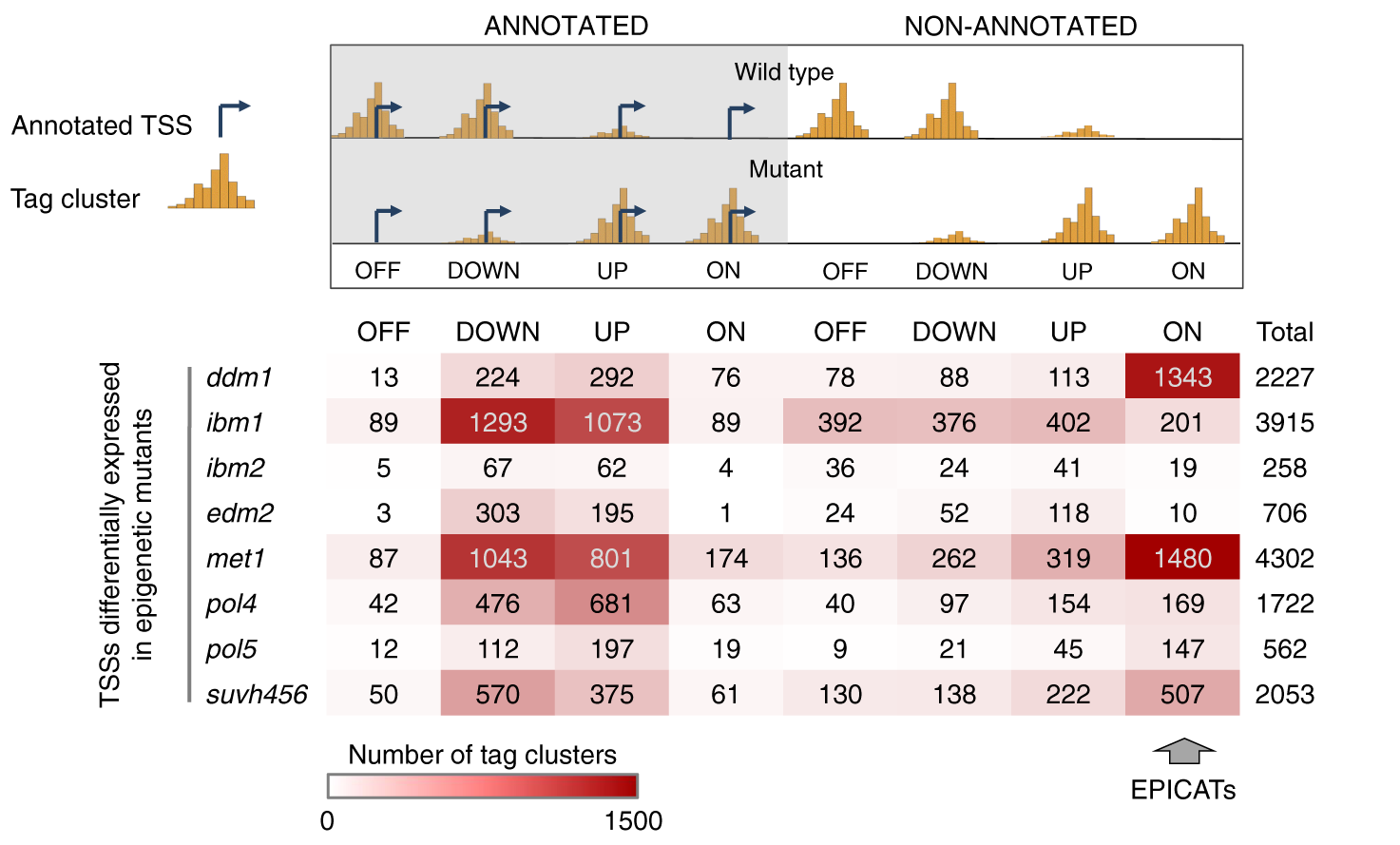
Figure 2: Classification of the TSSs differentially expressed in epigenetic mutants. A TSS was defined as ANNOTATED if the distance from its dominant CTSS to the nearest TAIR10-annotated TSS in the same orientation is less than 180 nt, or NON-ANNOTATED if not. NON-ANNOTATED TSSs activated de novo in the mutant backgrounds are named EPICATs (standing for EPigenetically Induced Consensus tAg clusTers).
4. Publications
4.1 Journals
1. Le NT, Harukawa Y, Miura S, Boer D, Kawabe A, Saze H. (2020) .Epigenetic regulation of spurious transcription initiation in Arabidopsis. Nat Commun. Jun 26;11(1):3224. doi: 10.1038/s41467-020-16951-w
2. Araki S, Le NT, Koizumi K, Villar-Briones A, Nonomura KI, Endo M, Inoue H, Saze H, Komiya R. (2020). miR2118-dependent U-rich phasiRNA production in rice anther wall development. Nat Commun. Jun 19;11(1):3115. doi: 10.1038/s41467-020-16637-3.
3. CREB-binding protein gene, HAC701, negatively regulates WRKY45-dependent immunity in rice. NA Espinas, TN Le, S Miura, Y Shimajiri, K Shirasu, H Saze. (2020) bioRxiv. doi: https://doi.org/10.1101/2020.08.26.268797
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
1. (Invited talk) Saze H. “Epigenetic regulation of intronic transposons in plant genomes”. Meeting: (Online) Annual meeting of the Botanical Society of Japan. Date and venue: 19 Sep. 2020, Nagoya, Japan.
2. (Invited online seminar) Saze H. “Epigenetic regulation of intragenic transposons and gene transcription in plant genomes.” Institution: I2BC External invited seminar, Institute for Integrative Biology of the Cell, CNRS, France. Date: 15 Jan. 2021.
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 Seminar Title in Full
- Seminar "Elucidation of molecular mechanisms regulating vessel cell differentiation through the control of activity of Arabidopsis VND7, a NAC transcription factor"
- Speaker: Dr. Risaku Hirai, Nara Institute Science and Technology
- Date and Location :Tuesday, October 13, 2020 - 15:00 to 16:00, C700, Lab3



