FY2021 Annual Report
Evolution, Cell Biology, and Symbiosis Unit
Assistant Professor Filip Husnik

Top row (from left to right): Yumiko Masukagami (+3), Filip Husnik, Dewi Langlet, Jinyeong Choi (+1). Bottom row: Iines Salonen, Daitian Xu, Mei Shimizu, Olivia Millar, Javier Tejeda Mora. Lab members not on the photo: Courtney Dunphy, Kimberlie Ward, Maria Eduarda Alves dos Santos, Machiko Shiomi, Pradeep Palanichamy.
Abstract
The Evolution, Cell Biology, and Symbiosis Unit studies the effects of symbiotic interactions on the evolution and cell biology of the involved symbiotic partners. We are especially interested in the role symbioses played in major evolutionary transitions such as the origin of the eukaryotic cell and its symbiotic organelles, mitochondria and plastids. However, we also study much younger symbioses such as those found in microbial eukaryotes and animals. Currently, our research projects mostly focus on symbiotic microorganisms and organelles found in single-celled eukaryotes (foraminifera, dinoflagellates, ciliates, and a few other lineages), plants (parasites and mycoheterotrophs), and animals (corals and insects).
1. Staff
- Dr. Dewi Langlet, Staff Scientist
- Dr. Courtney M. Dunphy, JSPS Postdoctoral Research Fellow
- Dr. Maria Eduarda Alves dos Santos, JSPS Postdoctoral Research Fellow
- Dr. Jinyeong Choi, NRF Korea Postdoctoral Research Fellow
- Dr. Yumiko Masukagami, Research Technician
- Dr. Iines Salonen, Visiting Researcher
- Kimberlie Ward, Technician
- Machiko Shiomi, Research Unit Administrator
- Javier Tejeda Moja, Ph.D. Student
- Pradeep Palanichamy, Ph.D. Student
- Yong Heng Phua, Research Intern, Rotation Student
- Mariia Naumova, Research Intern
- Zhuli Cheng, Rotation Student (Jan-Apr 2022)
- Rahel Ruppli, Rotation Student (Sep-Dec 2021)
- Daitian Xu, Research Intern (Dec 2021-March 2022)
- Olivia Millar, Research Intern (Oct 2021-March 2022)
- Mei Shimizu, Research Intern (Oct 2021-March 2022)
2. Collaborations
2.1 Endosymbioses of diverse microbial eukaryotes
- Researchers:
- Dr. Hidetaka Nomaki, JAMSTEC
- Dr. Iines Salonen, JAMSTEC
- Prof. Kazuhiko Fujita, University of Ryukyus
- Assoc. Prof. Naoya Shinzato, University of Ryukyus
- Assist. Prof. Kevin Wakeman, Hokkaido University
- Dr. Emma George, University of British Columbia
- Dr. Vittorio Boscaro, University of British Columbia
- Prof. Patrick Keeling, University of British Columbia
- Prof. Marek Elias, University of Ostrava
- Dr. Dovile Barcyte, University of Ostrava
2.2 Biology of coral symbioses
- Researchers:
- Dr. Masaru Mizuyama, AIST
- Dr. Akira Iguchi, AIST
- Assoc. Prof. James D. Reimer, University of Ryukyus
- Dr. 'Ale'a Dudoit, University of Hawaiʻi
- Assoc. Prof. Rob Toonen, University of Hawaiʻi
2.3 Cell biology and genomics of scale insect symbioses
- Researchers:
- Assist. Prof. Yu Matsuura, University of Ryukyus
- Dr. Bruno Humbel, OIST
- Dr. Hirotaka Tanaka, Ehime University
- Dr. Tomas Pluskal, IOCHB Prague
2.4 Eukaryogenesis and evolution of mitochondria
- Researchers:
- Dr. Akinori Yabuki, JAMSTEC
2.5 Genomics of parasitic and mycoheterotrophic plants
- Researchers:
- Assoc. Prof. Kenji Suetsugu, Kobe University
- Dr. Petra Svetlikova, OIST
3. Activities and Findings
3.1 Research Projects
3.1.1 Endosymbioses of diverse microbial eukaryotes
In this collection of smaller projects, we study endosymbioses of many diverse single-celled eukaryotes (reviewed in Husnik et al., Current Biology 2021; Figure 1) and develop tools for analyzing their genome evolution (Figure 2). During FY2021, in the projects led by unit members, we focused mostly on bacterial and algal symbioses of benthic dinoflagellates and foraminifera. Both of these protist groups are very rich in symbioses, yet extremely understudied. From benthic dinoflagellates, we selected toxin-producing lineages (Coolia, Amphidinium, Ostreopsis) as our model group where bacterial symbionts are sometimes hypothesized to be involved in toxin production. As a first step, we untangled their diversity, phylogeny, and in some cases microbial composition (Phua et al., Harmful Algae 2021; Phua et al., Phycologia 2022; Phua et al., submitted). Among foraminifera, we analyzed the algal symbiont diversity of large benthic foraminifera from coral reef flats and uncovered their extreme diversity and in most cases co-evolution with the foraminiferal hosts (Langlet et al., In prep.).
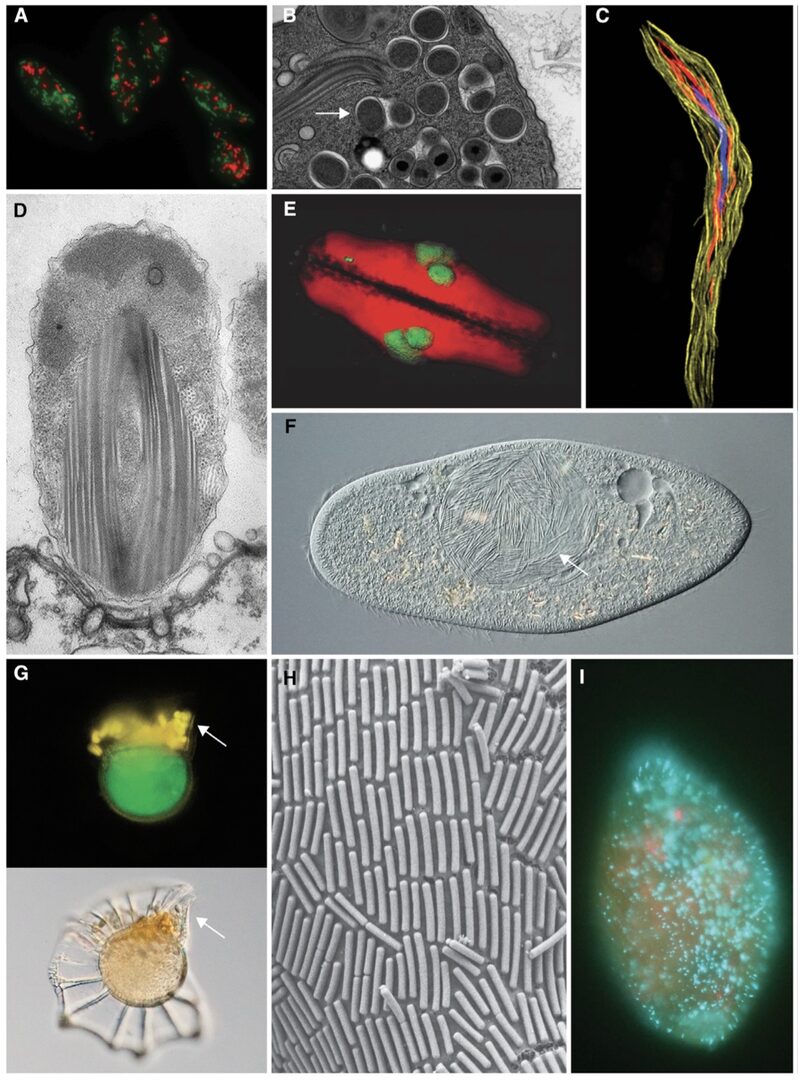
Figure 1: Examples of bacterial and archaeal symbioses with protists. Adopted from Husnik et al., Current Biology 2021.
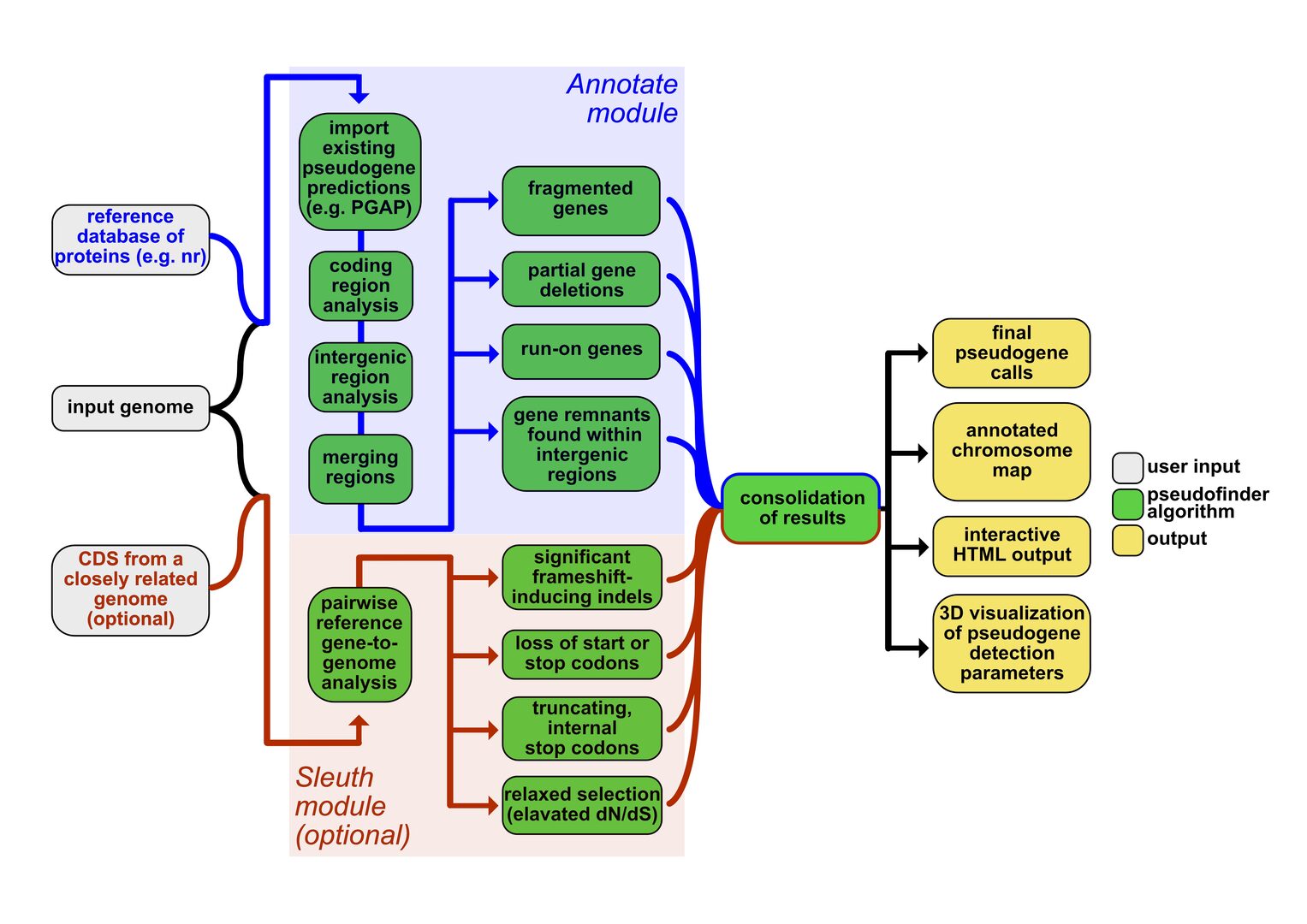
Figure 2. Pseudofinder is a newly developed software package for the identification of non-functional (pseudogenized) genes in bacterial and archaeal genomes. It was designed to automate genome annotation of bacterial symbionts that can often have more than half of all genes pseudogenized. However, it is widely applicable to any prokaryotic genomes. The figure shows an overview of its workflow. For more information, please visit its bioRxiv preprint or GitHub page.
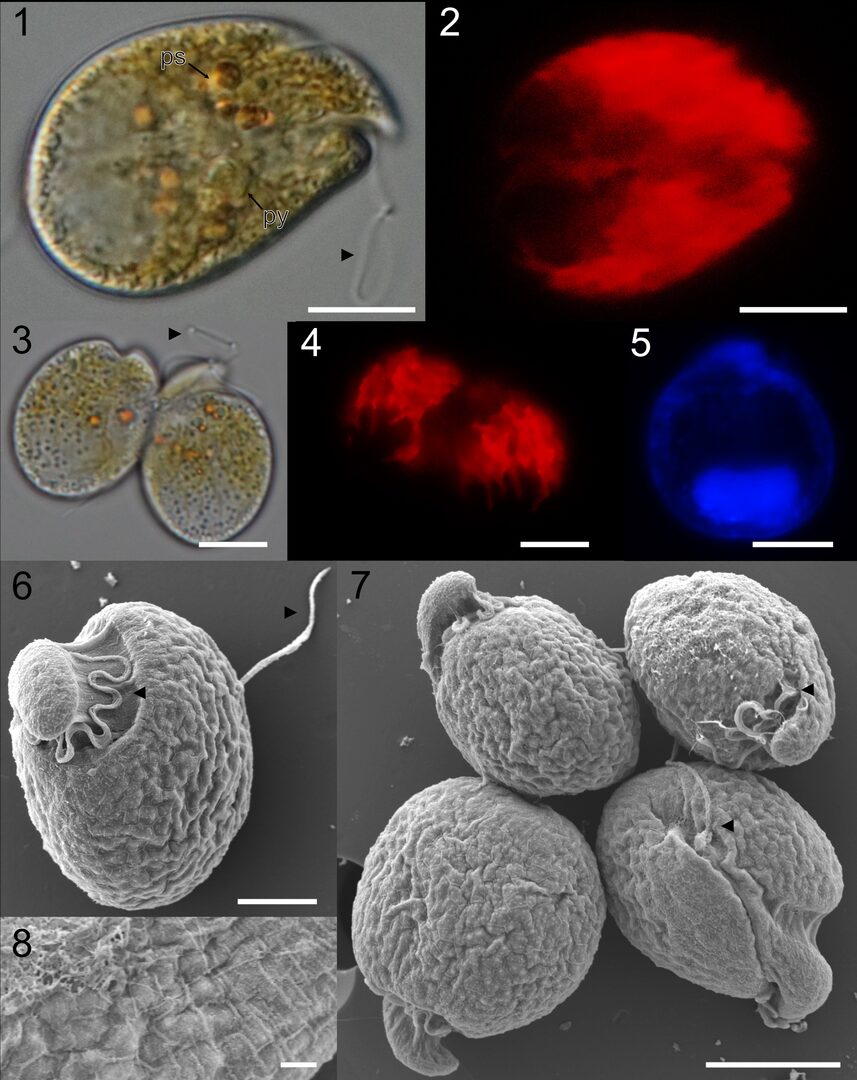
Figure 3: Benthic dinoflagellates studied in the unit include mainly toxic genera Amphidinium, Coolia, and Ostreopsis. On the image plate, Amphidinium pagoensis sp. nov. was characterized with light, fluorescent, and scanning electron micrographs (SEM). Adopted from Phua et al., Phycologia 2022.

Figure 4: Large benthic foraminifera used in our unit as model species for eukaryote-eukaryote symbioses. Top left to bottom right: Amphisorus sp. (houses dinoflagellate symbionts), Peneroplis sp. (houses red-algal symbionts), Baculogypsina sphaerulata, Calcarina gaudichaudii, and Amphistegina lobifera (all three house diatom symbionts). These foraminiferal species are a major component of sand beaches. Their calcium carbonate shells are famous in the Ryukyu Islands as 'star and sun sand' (hoshizuna). Images from Okinawa by Dewi Langlet.
3.1.2 Coral symbioses
In this project, we investigate the diversity and functions of microbial symbionts associated with coral animals. During FY2021, we created a global data set of coral DNA samples and used amplicon and shotgun metagenome sequencing to reveal the distribution of their associated bacteria and eukaryotes across regions with different seawater conditions. Our results show that Palythoa corals are associated with dinoflagellate species of the genera Cladocopium, Durusdinium and Symbiodinium (Santos et al., In prep; Figure 5). Coral samples from the Atlantic and Indo-Pacific differ in their dinoflagellate composition. Stress-tolerant symbionts (e.g., Durusdium) were found associated with corals from the Red Sea and Iotorishima Island, Japan (warmest seawater temperature and low pH seawater conditions, respectively). Among bacterial genera, the most common was the likely mutualistic Endozoicomonas (Figure 6). Interestingly, some coral species also hosted likely pathogenic bacteria from the Vibrio genus.
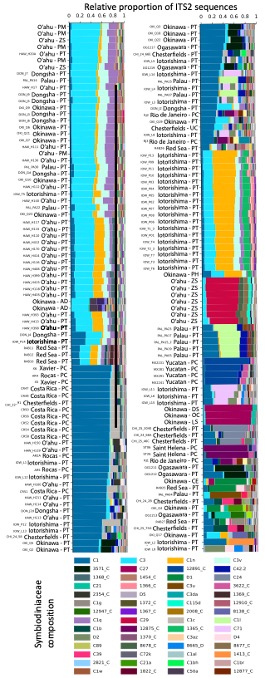
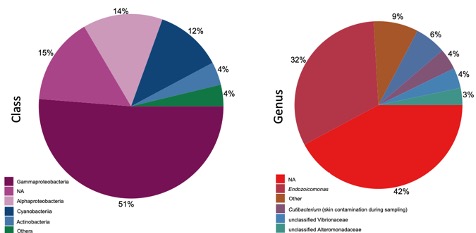
Figure 6: Relative abundance of the bacterial diversity found in all coral samples. The diversity was estimated using amplicon sequencing of the 16S rRNA gene.
Figure 5 (on the left): Relative proportion of dinoflagellates (family Symbiodiniaceae) associated with each coral sample. Symbiodiniaceae composition was estimated using amplicon sequencing of the ITS2 gene. Only the most abundant (>0.01% of all reads) sequences are displayed. Labels indicate the Symportal framework database ID number followed by the genus the sequence is from (C=Cladocopium and D=Durusdinium). Host species codes are PT (Palythoa tuberculosa) and PC (P. caribaeorum), in addition to outgroups PM (P. mutuki), PH (P. heliodiscus), AD (Acropora digitifera), CE (Ctenactis echinata), DS (Danafungia scraposa), LS (Lytophyllon scabra), OC (Pobadacia crustacea), UC (Umimayanthus chanpuru), ZS (Zoanthus sansibaricus).
3.1.3 Cell biology and genomics of scale insect symbioses
During this FY, we sequenced and analyzed metagenomes from over 40 scale insect species and reconstructed the evolution of symbioses in this insect group (Choi and Lee, Systematic Entomology 2022; Choi et al., In prep.). For selected species maintained in our unit as laboratory colonies, we used electron microscopy, confocal microscopy, microCT, and metabolomics to understand the functional role of the symbioses in detail.
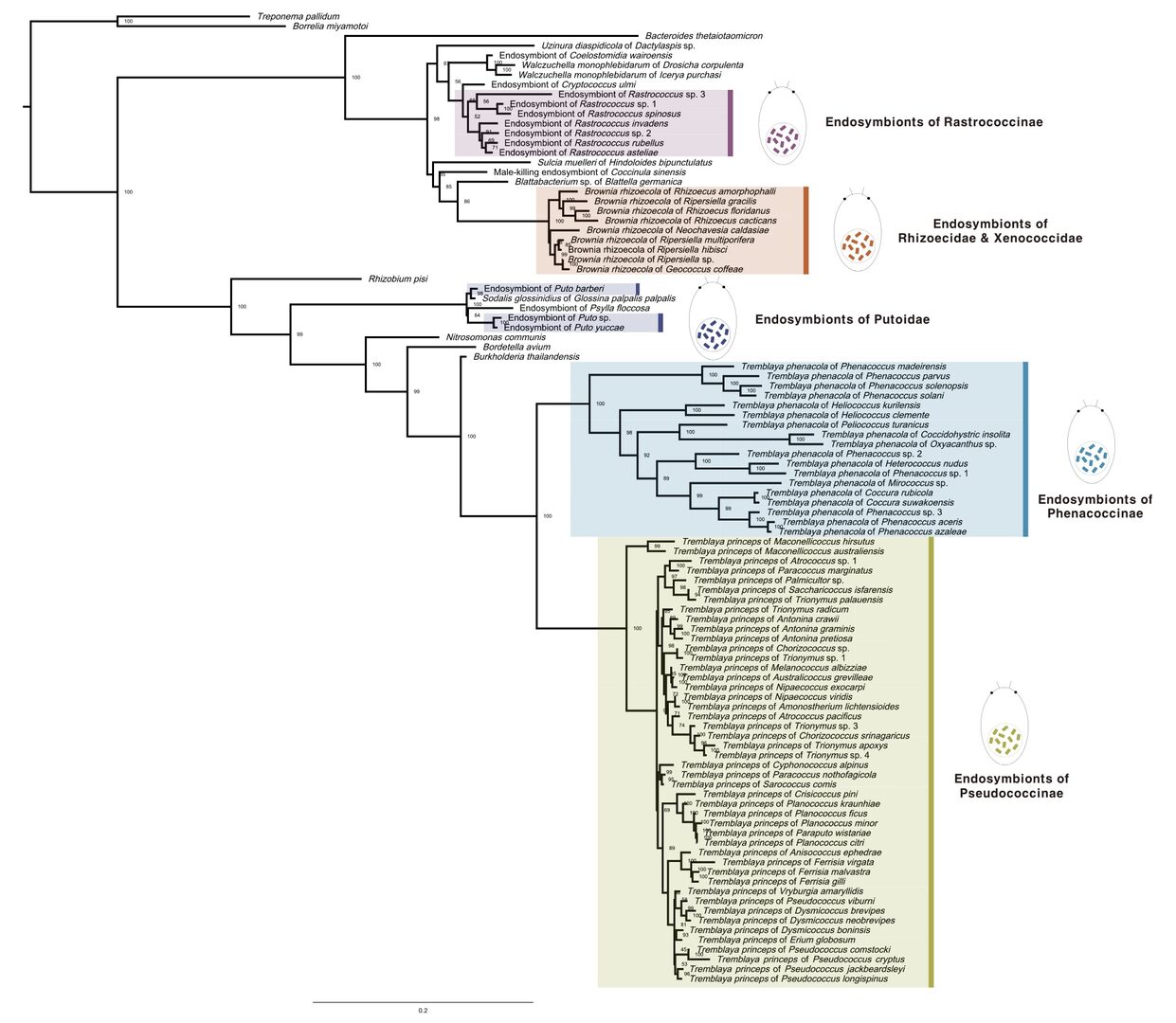
Figure 7: Phylogenetic tree of mealybug symbionts. Adopted from Choi and Lee, Systematic Entomology 2022.
Figure 8: Cottony cushion scale (Icerya purchasi, Monophlebidae, Coccomorpha) is one of a few insect species that is a hermaphrodite. Our group studies its bacterial symbionts. Image from Okinawa by Jinyeong Choi.
3.1.4 Eukaryogenesis and evolution of mitochondria
In this mostly experimental project, we focus on the evolution of heterotrophic (predatory) protists and protists with gene-rich mitochondrial genomes. During this FY, several species were selected as laboratory models, and we successfully established multiple novel methods for studying even the most challenging eukaryotes (e.g., cells smaller than one micrometer). Among other methods, we optimized fluorescent labeling of their diverse cell compartments, cell enrichment via FACS, their bacterial prey characterization, and single-cell genomics, transcriptomics, and imaging.
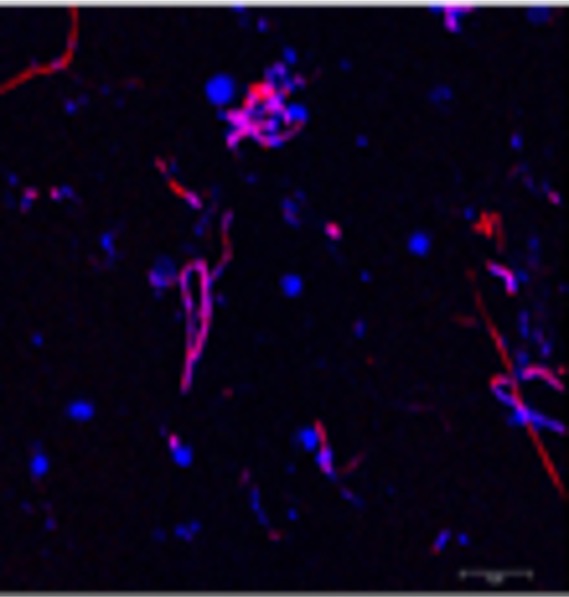
Figure 9: Super-resolution image of the nanoeukaryote Symbiomonas scintillans with its bacterial prey. Large circular nuclei of Symbiomonas are stained in blue with DAPI. Two bacterial lineages are stained in red and magenta with fluorescently labeled 16S rRNA probes (FISH).
3.1.5 Genomics of parasitic and mycoheterotrophic plants
In this collaborative project with prof. Kenji Suetsugu (Kobe University), we provide our expertise in eukaryotic genomics and plastid evolution. For example, Balanophora species (B. fungosa; Figure 10) are parasitic plants with extremely reduced plastid genomes which are also the most AT-rich plastid genomes reported so far (Figure 9). During this FY, we sequenced and analyzed plastid genomes of 14 parasitic and mycoheterotrophic plants from Okinawa and mainland Japan (Svetlikova et al., In prep.).
Figure 10. Balanophora fungosa from Okinawa. Image by Petra Svetlikova.
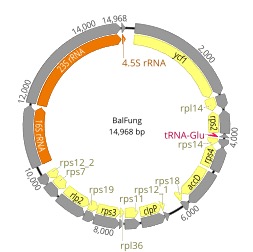
Figure 11. Highly reduced plastid genome of the non-photosynthetic holoparasite B. fungosa.
4. Publications
4.1 Journals
Syberg-Olsen MJ, Graber AI, Keeling PJ, McCutcheon JP, Husnik F: Pseudofinder: detection of pseudogenes in prokaryotic genomes [https://github.com/filip-husnik/pseudofinder], bioRxiv 2021, doi: 10.1101/2021.10.07.463580.
Giannotti D, Boscaro V, Husnik F, Vannini C, Keeling PJ: The ‘other’ Rickettsiales: an overview of the family ‘Candidatus Midichloriaceae’, Applied and Environmental Microbiology 2022, https://doi.org/10.1128/aem.02432-21.
Phua YH, Husnik F, Lemer S, Wakeman KC: Phylogeny of benthic dinoflagellate Amphidinum (Dinophyceae) from the South Pacific Islands of Guam and Okinawa, with the description of A. pagoensis sp. nov. and A. uduigamensis sp. nov., Phycologia 2022, https://doi.org/10.1080/00318884.2022.2033948.
Choi J, Seunghwan L. Higher classification of mealybugs (Hemiptera: Coccomorpha) inferred from molecular phylogeny and their endosymbionts. Systematic Entomology 2022, https://doi.org/10.1111/syen.12534.
Fontanier C, Deflandre B, Rigaud S, Mamo B, Dubosq N, Lamarque B, Langlet D, Schmidt S, Lebleu P, Poirier D, Cordier MA, Grémare A. Live (stained) benthic foraminifera from the West-Gironde Mud Patch (Bay of Biscay, NE Atlantic): Assessing the reliability of bio-indicators in a complex shelf sedimentary unit. Continental Shelf Research 2022, 232: 104616.
Kise H, Montenegro J, Santos MEA, Hoeksema BW, Ekins M, Ise Y, Higashiji T, Fernandez- Silva I, Reimer JD. Evolution and phylogeny of glass-sponge- associated zoantharians, with description of two new genera and three new species. Zoological Journal of the Linnean Society 2022, 194(1): 323-347.
Soares MO, Kitahara MV, Santos MEA, Bejarano S, Rabelo EF, Cruz ICS. The flourishing and vulnerabilities of zoantharians on Southwestern Atlantic reefs. Marine Environmnetal Research 2021 173, 105535.
Husnik F, Tashyreva D, Boscaro V, George EE, Lukes J, Keeling PJ: Bacterial and archaeal symbioses with protists. Current Biology 2021, 31(13): PR862-R877.
Salonen IS, Chronopoulou P-M, Nomaki H, Langlet D, Tsuchiya M, Koho KA: 16S rRNA gene metabarcoding indicates species-characteristic microbiomes in deep-sea benthic foraminifera. Front. Microbiol. 2021, https://doi.org/10.3389/fmicb.2021.694406.
Phua YH, Roy MC, Lemer S, Husnik F, Wakeman KC: Diversity and toxicity of Pacific strains of the benthic dinoflagellate Coolia (Dinophyceae), with a look at the Coolia canariensis species complex. Harmful Algae 2021, https://doi.org/10.1016/j.hal.2021.102120.
4.2 Books and other one-time publications
Nothing to report.
4.3 Oral and Poster Presentations
- Langlet, D., Husnik, F. The diversity and role of foraminiferal symbionts in the benthic ecosystem functioning in subtropical mudflats, TMS Foraminifera Festival, Online, August (2021).
- Husnik, F. The DOE JGI Symposium ‘From New Lineages of Life to New Functions’, Invited Talk, Online, October 14 (2021).
- Husnik, F. The 34th Annual Meeting of the Japanese Society of Microbial Ecology, Invited Talk, Online, November 1 (2011).
- Husnik, F. The 4th Asian Congress of Protistology, Invited Talk, Online, November 11 (2021).
- Langlet, D., Ruppli, R., Fujita, K., Husnik, F. Metagenomics reveals diverse host-specific photosymbionts in Okinawan large benthic foraminifera, OIST-Tohoku U. Joint Symposium, Poster, OIST, Okinawa, April 25-26 (2022).
- Masukagami, Y., Millar, O., Naumova, M., Husnik, F. Developing experimental methods for the tiniest tiny marine predators and their symbioses. OIST-Tohoku U. Joint Symposium, Poster, OIST, Okinawa, April 25-26 (2022).
- Santos, M.E.A., Reimer, J.D, Mizuyama, M., Kise, H., Boo, W. H., Iguchi, A. Dudoi, A., Toonen, R., Kitahara, M.V., Husnik, F. Global diversity of coral endosymbionts. OIST-Tohoku U. Joint Symposium, Poster, OIST, Okinawa, April 25-26 (2022).
5. Intellectual Property Rights and Other Specific Achievements
External funding acquired by unit members:
- NRF Korea Postdoctoral Research Fellowship (Jinyeong Choi)
- JSPS Postdoctoral Research Fellowship (Maria Eduarda Alves dos Santos, from September 2021)
- JSPS Postdoctoral Research Fellowship (Courtney M. Dunphy, from December 2020)
- JSPS Grant-in-Aid for Early-Career Scientists (main PI Yumiko Masukagami; FYs 2021-2023)
- JSPS Grant-in-Aid for Scientific Research B (F. Husnik co-PI; FYs 2021-2023)
- JSPS Grant-in-Aid for Early-Career Scientists (main PI F. Husnik; FYs 2021-2022)
- Aure Hawaii-Okinawa grant (Maria Eduarda Alves dos Santos and F. Husnik co-PIs; FY 2020, FY 2021)
- JSPS Grant-in-Aid for Research Activity Start-up (main PI F. Husnik; FYs 2020-2021)
6. Meetings and Events
6.1 Seminar: Biology of apicomplexans and dinoflagellates
- Date: August 19, 2021
- Venue: OIST Campus Lab4
- Speaker: Assist. Prof. Kevin Wakeman (Hokkaido University)
6.2 Seminar: Endosymbioses of anaerobic protists
- Date: October 7, 2021
- Venue: OIST Campus Lab4
- Speaker: Assoc. Prof. Naoya Shinzato (University of Ryukyus)
6.2 Seminar: Symbioses of hemipteran insects
- Date: November 18, 2021
- Venue: OIST Campus Lab4
- Speaker: Assist Prof. Yu Matsuura (University of Ryukyus)
7. Other
Masukagami, Y. - Kyuyou high school, Career talk, Kyuyou High School, Okinawa, Japan, 10. Nov. 2021.
Masukagami, Y., Akamine, Y., Takahashi, A. - HiSci lab, Female Scientist Career Talk, Onna Village Fureai Experience Learning Center, Okinawa, Japan, 13. Mar. 2021.